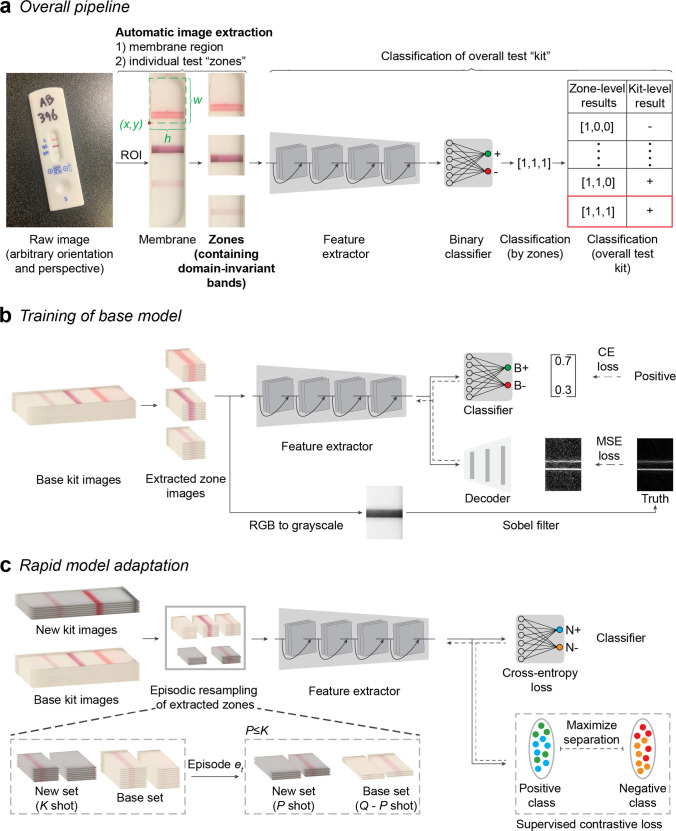
Fig. 2. Overview of AutoAdapt POC machine-learning pipeline.
a From a raw input image of an assay kit, a correction of orientation and perspective is applied to segment an image of an assay kit. From the assay kit image, a segmentation model based on Mask R-CNN is used to extract the membrane region of interest (RoI). Based on measured kit-specific parameters (details in Supplementary Table 3), individual zones are cropped, and passed through a software pipeline consisting of a feature extractor followed by a binary classifier. Classification of each zone allows, via a kit-specific lookup table, for a final classification of assay result (kit-level classification or result) as positive, negative, or invalid. b The feature extractor is pre-trained on the base kit using self-supervised learning task over edge-filtered patterns and fully-supervised binary classification task. For each zone, fully-supervised binary classification is carried out with cross-entropy loss with the annotated binary labels. Sobel filter is used to highlight the edge pixels between the band and the background of the membrane. The edge image after normalization is used as ground truth and the learning process is used to reconstruct an image that resembles the ground truth edge image, with the quality measured in MSE (Mean Square Error). The solid and dashed arrows indicate forward processing and gradient backpropagation, respectively, during the learning process. c Model adaptation is carried out by supervised contrastive learning to regularize the feature extractor and fully-supervised learning to learn an adapted classifier for the new kit. A sampling strategy to build an episode with Q (e.g., 32) images per class is used: for each class (positive or negative), given K (e.g., 10) images available, P (e.g., 4) images are subsampled from the new kit and mixed with Q-P images of the base kit.