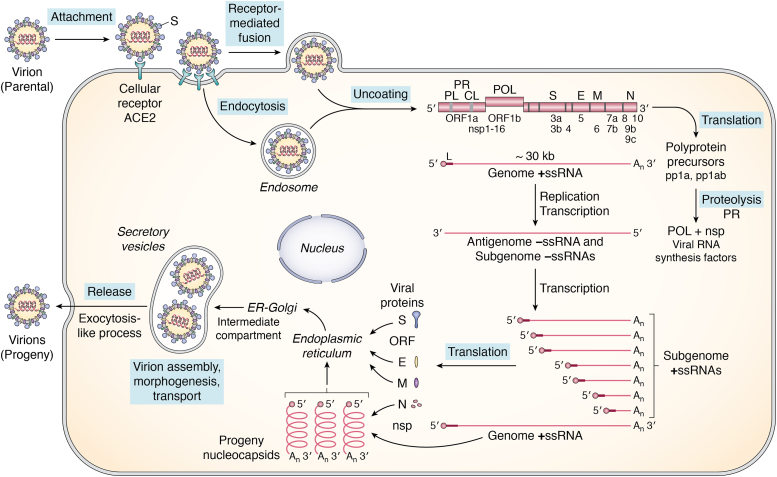
Figure 1.
Schematic diagram summary of the SARS-CoV-2 coronavirus multiplication cycle. SARS-CoV-2 virus possesses a positive-sense, single-stranded RNA genome of ∼30-kb. Virus multiplication occurs in the cytoplasm. Following virion attachment by binding of the viral S spike protein to the cellular ACE2 receptor protein, entry occurs by receptor-mediated fusion of the viral envelope with the host plasma membrane, releasing the ribonucleocapsid complex containing genome +ssRNA into the cytoplasm. The 5′-capped genome RNA functions as mRNA and is translated directly by the host cell protein-synthesizing machinery to generate two polyprotein precursor products, pp1a from ORF1a, and pp1ab from ORF1a and ORF1b following a −1 ribosomal frameshift to read frame1b. The polyprotein precursors undergo autoproteolytic processing by viral PL and CL proteases to generate nonstructural proteins (nsp 1–16) that include the viral RNA polymerase and factors required for viral RNA biogenesis. The parental genome +ssRNA next functions as a template for replication and transcription to produce full-length antigenomic –ssRNA (that is the template for synthesis of full length +ssRNA progeny genomes) and a set of subgenomic –ssRNAs (that are the templates for subgenomic +ssRNAs that are the mRNAs for synthesis of virion structural proteins N, M, E and S as well as several ORF accessory proteins that affect the host response to infection). Viral RNA synthesis occurs in association with cellular membrane structures, as does progeny virion assembly and morphogenesis followed by release by an exocytosis-like process.