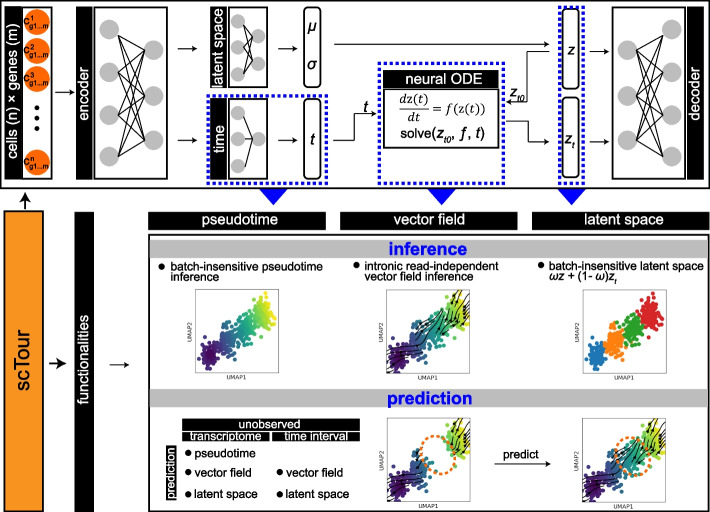
Fig. 1.
scTour framework. With a gene expression matrix as input, two encoder networks are used to both generate the distribution parameters of the approximate posterior (latent space, ) and assign a time point to each cell (time, ). The sample from the posterior at the initial state () along with the times () of cells are input into a neural ODE to yield another series of latent representations . A decoder network then reconstructs the input using the latent and . This model can be used to infer the developmental pseudotime, transcriptomic vector field, and latent representations of cells in an unsupervised manner, as well as to predict the cellular dynamics of unobserved transcriptomes or time intervals