Figure 2.
Deletion of SETD7 does not influence the pluripotency of hESCs
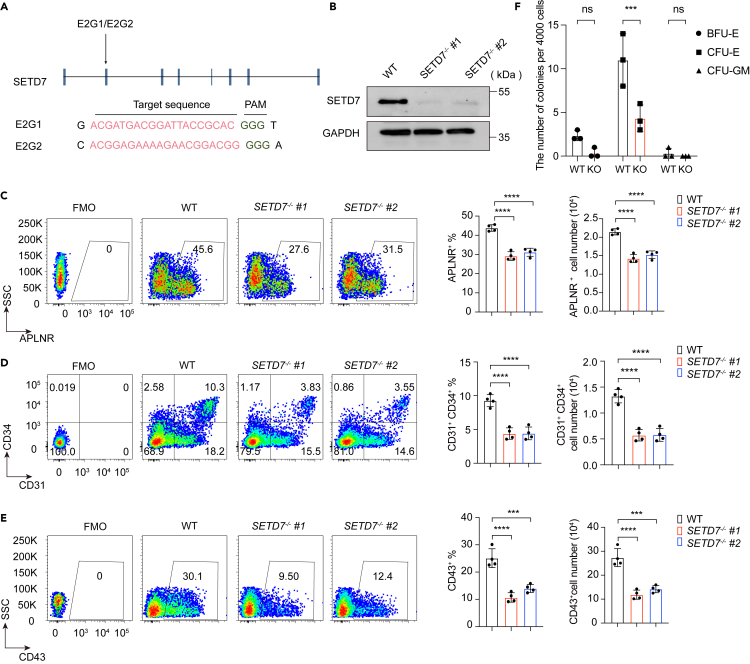
(A) Schematic illustration of the strategy to target SETD7 exon 2 by CRISPR/Cas9 technology. E2G1 and E2G2 refer to two sgRNAs with targeting sequences (red) and protospacer adjacent motifs (PAMs) (green).
(B) Western blot showing the expression of SETD7 in WT, SETD7−/− #1 and #2 cell lines. GAPDH served as the loading control.
(C–E) Representative flow cytometry plots (left) and bar graphs showing the proportion (middle) and the cell number (right) of APLNR+ LPM cells at day 2 (C), CD31+CD34+ EPCs at day 5 (D) and CD43+ HPCs (E) at day 8 in WT, SETD7−/− #1 and SETD7−/− #2 cells during hematopoietic differentiation. (n = 4, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, ordinary one-way ANOVA). Data are means ± SD. (F) The number of colonies (BFU-E, CFU-E and CFU-GM) generated from day 12 differentiated HPCs of WT or SETD7−/− cells. (n = 3, ∗∗∗p < 0.001, ns, no significance, ordinary one-way ANOVA). Data are means ± SD. See also Figure S1 and Table S2.