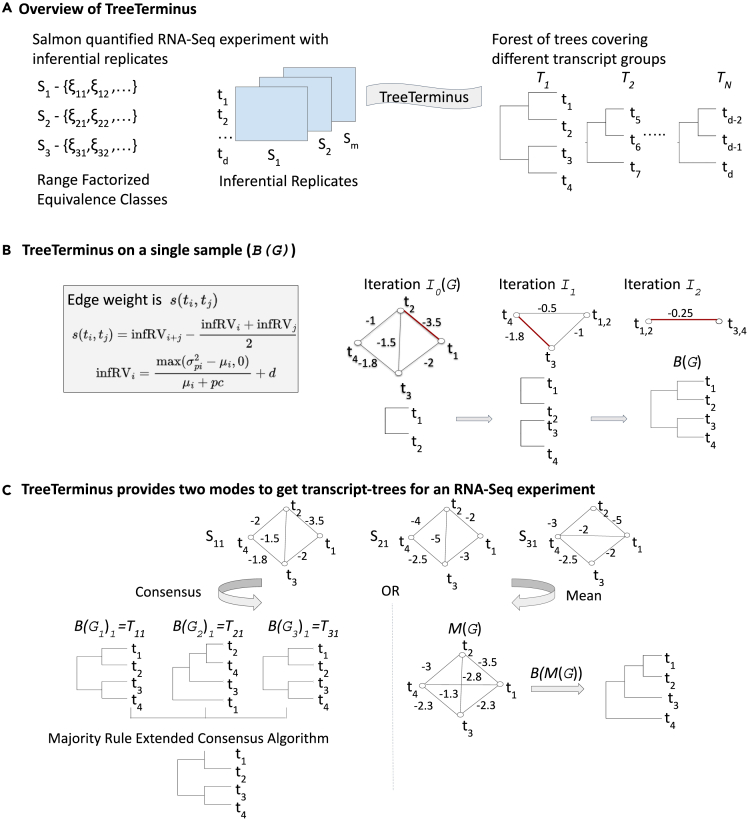
Figure 1.
Schematic overview of TreeTerminus pipeline
(A) Taking an RNA-Seq experiment as an input, that has been quantified with Salmon and for which inferential replicates have been generated, TreeTerminus outputs a forest of trees.
(B) Toy example demonstrating how from the transcript graph (G), how a forest shall be generated by TreeTerminus for a single sample using the procedure B(G). A forest will consist of multiple trees when the graph consists of disconnected components or some of the necessary conditions that are required for aggregating transcripts/groups are not met. The color red indicates the edge with the lowest weight chosen for aggregation at each iteration.
(C) When there are multiple samples—TreeTerminus provides two modes to generate the forest, Mean and Consensus. In the toy example, both procedures are demonstrated using a single transcript group, thus the forest consists of a single tree.