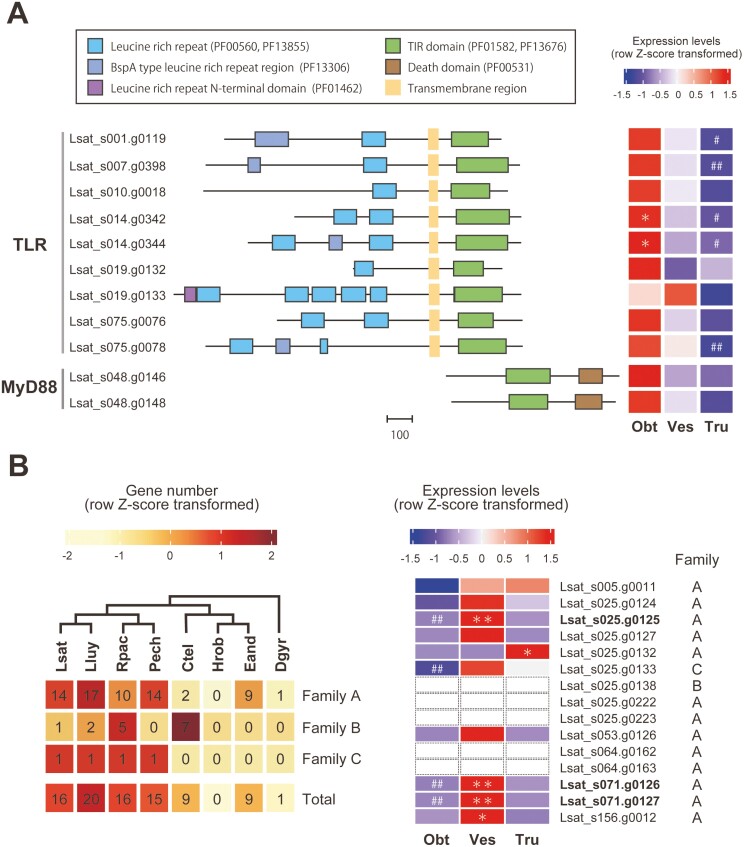
Figure 3.
Toll-like receptor, MyD88, and lysozyme genes in the L. satsuma genome. (A) TLR and MyD88. Domain organizations are shown on the left. The scale bar indicates the number of amino acid residues. A heat map of relative gene expression levels (row Z-score transformed) is shown on the right. (B) Lysozymes. Gene numbers of OGs containing putative lysozyme genes (family A, B, and C) and their totals are shown on the left. The cladogram at the top is based on phylogenetic relationships inferred from the present study (Supplementary Fig. S1). All L. satsuma genes in these OGs were annotated as lysozymes by a BLAST search against the SwissProt database, and all except one, possessed a destabilase domain (PF05497). Heat map of relative expression levels (row Z-score transformed) of lysozyme genes with a destabilase domain (PF05497), including three vestimentum-specific DEGs are shown on the right. Expression of Lsat_s025.g0138, 0222, g0223, Lsat_s064.g0162, g0163 were not detected in any of the three regions. Lsat, L. satsuma; Lluy, L. luymesi; Rpac, R. pachyptila; Pech, P. echinospica; Ctel, C. teleta; Hrob, H. robusta; Eand, E. andrei; Dgyr, D. gyrociliatus; Obt, obturacular region; Ves, vestimental region; Tru, trunk region; * or **, genes with expression levels significantly higher in one region than other regions (FDR < 0.05 or 0.01, respectively); # or ##, genes with expression levels significantly lower in a specific region than in other regions (FDR < 0.05 or 0.01, respectively).