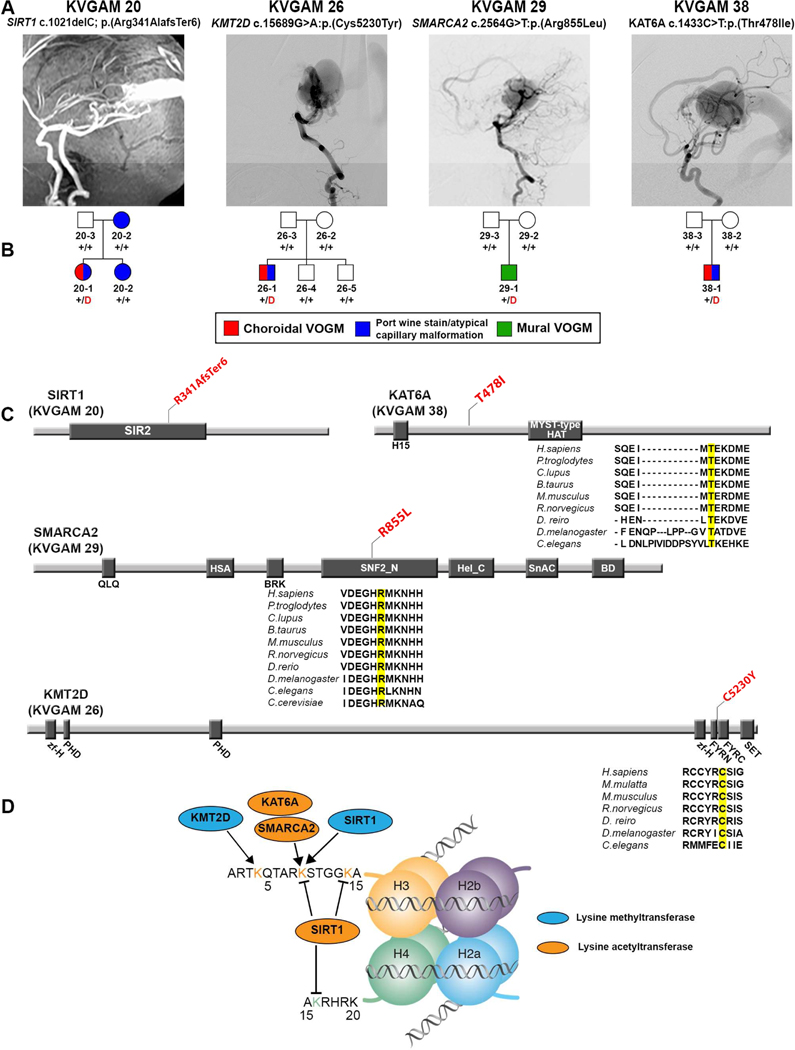
Figure 2. Damaging de novo mutations in chromatin modifiers in VOGM.
(A) Magnetic resonance angiographies and a digital subtraction angiography reconstruction demonstrating VOGM in probands from four pedigrees.
(B) Pedigree structures of VOGM kindreds. For each kindred, the gene and mutation, the angiographic image, and pedigree structure are shown. Subjects with atypical capillary malformations are denoted by blue symbols. Red ‘D’ denotes damaging mutation, ‘+’ denotes wild type sequence.
(C) Linear representation of functional domains of SIRT1, KMT2D, SMARCA2, and KAT6A, with location of VOGM mutations. Functional domains are represented by dark rectangles. Amino acid changes (red) are located on the protein structure. For missense mutations, phylogenetic conservation of the wild-type amino acid is shown, with the mutated amino acid in yellow. SIR2 = Sirtuin catalytic domain, SIR2 Domain; PHD = Zinc Finger PHD type; MOZ_SAS = Histone acetyltransferase domain, MYST-type; zf-H = PHD-like zinc binding domain; FYRN = F/Y-rich domain - F/Y-rich N-terminus motif; FYRC = F/Y-rich domain - F/Y-rich C-terminus motif; SET = Su(var)3–9, Enhancer-of-zeste and Trithorax; QLQ = Glutamine-Leucine-Glutamine domain; HAS = Helicase-SANT associated domain; BRK = BRK domain; SNF2_N = SNF2-related, N-terminal domain; Hel_C = Helicase C-terminal domain; SnAC = Snf2-ATP coupling, chromatin remodeling complex; BD = Bromodomain.
(D) Schematic of histone mark modifications by SIRT1, KMT2D, SMARCA2, and KAT6A.