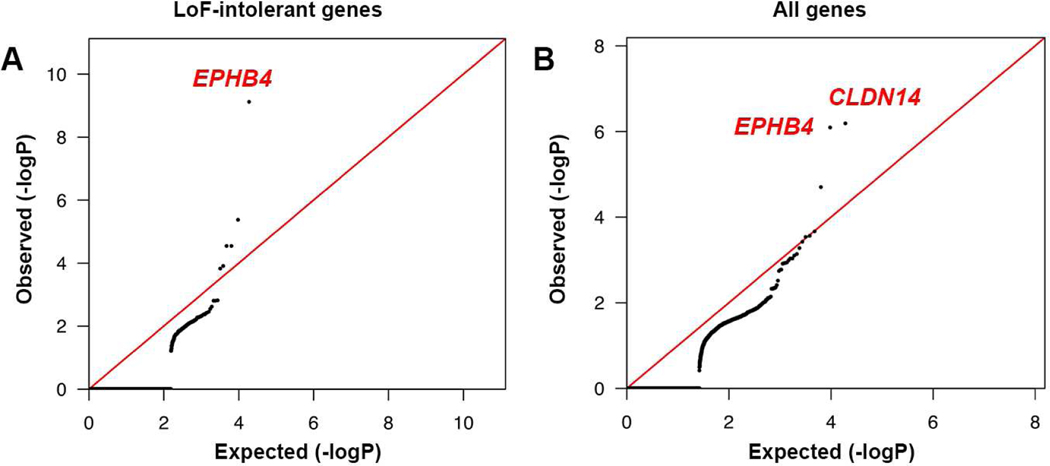
Figure 3. Exome-wide significant enrichment of rare damaging transmitted mutations in EPHB4 and CLDN14.
(A) Quantile-quantile plots of observed vs. expected binomial test p-values for rare damaging (D-mis+LoF) variants with MAF ≤ 2 × 10−5 in the Genome Aggregation database (gnomAD) in LoF-intolerant genes (pLI ≥ 0.9).
(B) Quantile-quantile plots of observed vs. expected binomial test p-values for rare damaging (D-mis+LoF) variants with MAF ≤ 2 × 10−5 in gnomAD for all genes. MAF = Minor allele frequency; D-mis = Missense mutations predicted to be deleterious per MetaSVM; LoF = Canonical loss-of-function mutations (stop-gains, stop-losses, frameshift insertions or deletions, and canonical splice site mutations).