Figure 4. YAP1 and TAZ regulate inflammatory signals through TEAD-independent gene networks.
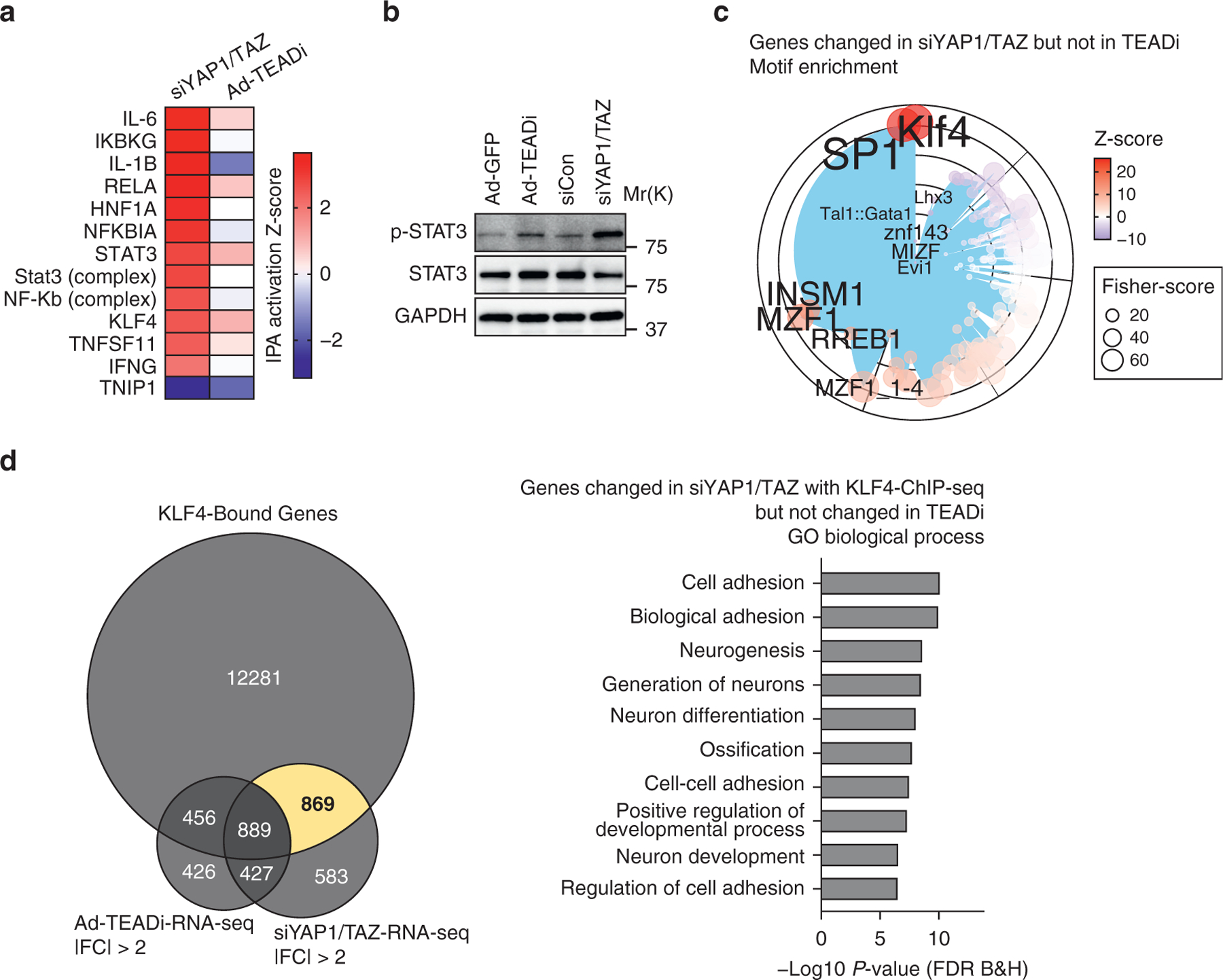
(a) Heatmap showing the activation Z-score for selected IPA upstream transcriptional regulators for genes differentially regulated in siYAP1/TAZ or TEADi datasets. (b) Western blot showing the expression level of p- and total STAT3 in BCC cells treated with the indicated conditions. (c) Graph showing the transcription factor binding site enrichment analysis in genes exclusively regulated by siYAP1/TAZ; only the top enriched transcription factors are highlighted; color represents Z-score, and size represents Fisher score. (d) Venn diagram indicating the overlap between KLF4 ChIP-seq-target genes and genes differentially regulated by siYAP1/TAZ and TEADi and selected GO biological process terms from the area highlighted in yellow. Ad-GFP, adenoviruses expressing GFP; Ad-TEADi, adenoviruses expressing a GFP-tagged TEAD inhibitor; BCC, basal cell carcinoma; ChIP-seq, chromatin immunoprecipitation sequencing; FC, fold change; FDR, false discovery rate; GO, gene ontology; IPA, ingenuity pathway analysis; p-, phosphorylated; RNA-seq, RNA sequencing; siCon, small interfering RNAs targeting control; siYAP1/TAZ, small interfering RNAs targeting YAP1 and TAZ; TEADi, TEAD inhibitor
