Fig. 3.
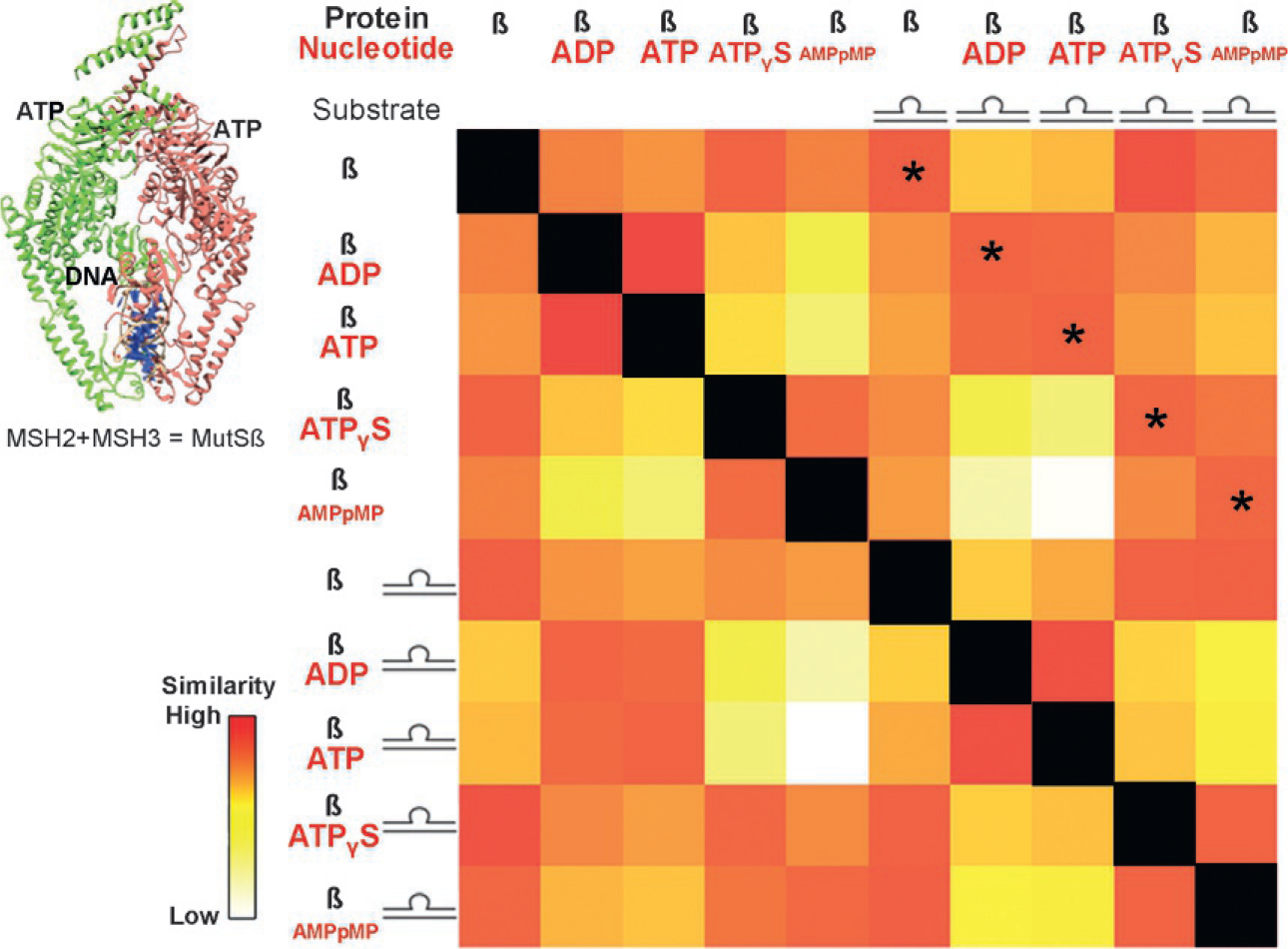
Conformational changes of MutSβ due to substrate binding. MutSβ aids in repairing DNA loops and is powered by ATP hydrolysis. SAXS collected on MutSβ with and without DNA and with nucleotides ADP, ATP, and non-hydrolysable analogues AMPpMP and ATPγS were compared against one another in an SSM. The asterisked diagonal compares measurements with and without DNA in the presence of the same nucleotide. The indicated similarity across this diagonal highlights that, mechanistically, the nucleotide state determines the conformation of the protein, not DNA binding.
