Figure 1:
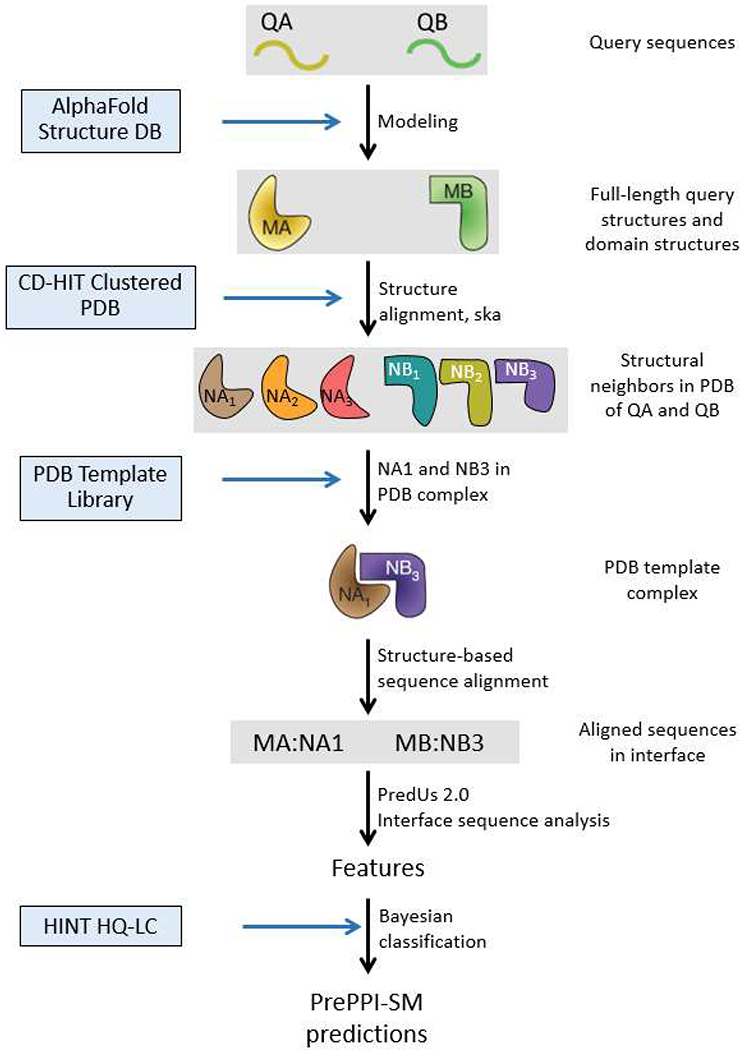
PrePPI’s structural modeling (SM) pipeline: Structures for query proteins, QA and QB, are taken from the AlphaFold Protein Structure Database [13] and parsed into domains with definitions from the Conserved Domain Database (CDD) [22]. Structural neighbors in the PDB [3] for full length protein and domain structures are obtained from the ska structural alignment program [31]. If structural neighbors of two query proteins appear together in a PDB complex, this structure defines a template, NA1:NB3, used to create a structure-based sequence alignment with which an interface for the query proteins, QA:QB, is evaluated based on the overlap of the query and template residues [1]. The interaction is then scored based on a number of features [1, 2] and trained on the HINT HQ-LC database [10], as the positive set, and a negative set described in Methods to produce a fully connected Bayesian network used to evaluate the model.
