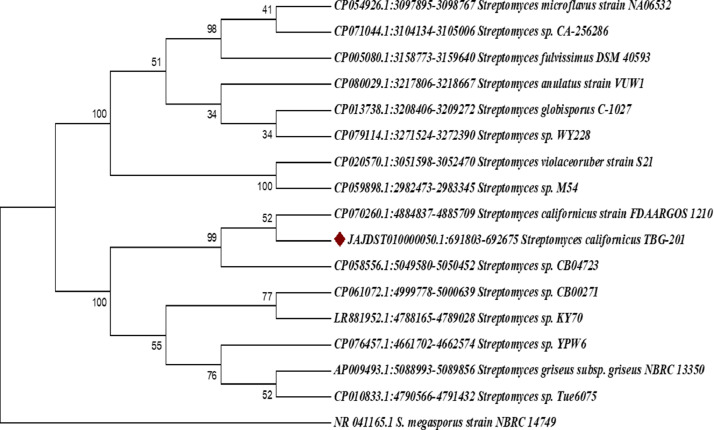
Fig. 1.
Evolutionary relationships of taxa of S. californicus TBG-201 based on 16S rDNA. Using the Neighbor-Joining method, the evolutionary history was deduced [4]. The bootstrap consensus tree inferred from 1000 replicates represents the evolutionary history. Next to each branch is the percentage of replicate trees in which the related taxa grouped together in the bootstrap test (1000 replicates) [5]. The evolutionary distances were calculated using the Jukes-Cantor method [6] and are in the units of the number of base substitutions per site. The analysis involved 17 nucleotide sequences. Codon positions included were 1st+2nd+3rd and noncoding. For each sequence pair, the ambiguous positions were eliminated. There were a total of 1474 positions in the final dataset.