Extended Data Fig. 8 |. T cell deletion of Ar.
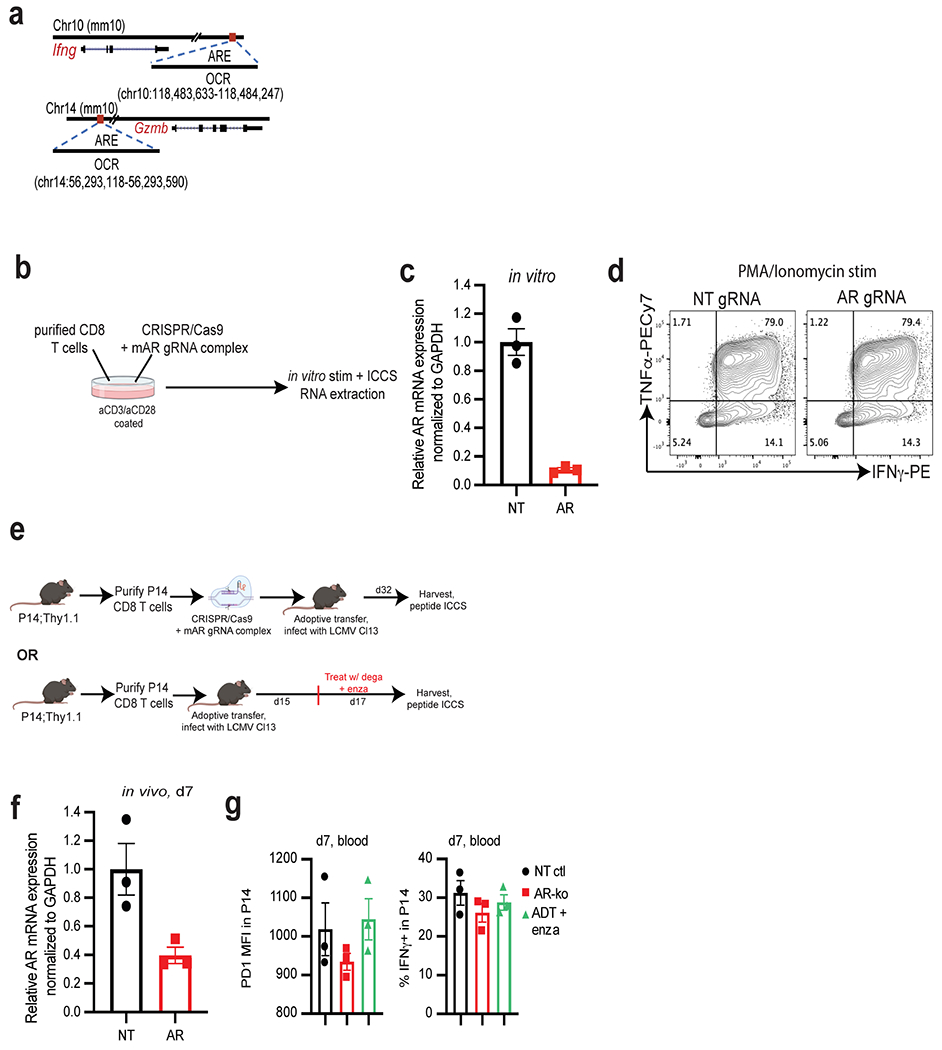
a, Open chromatin regions (OCRs) containing predicted androgen receptor elements (AREs) in Ifng and Gzmb loci. b, Experimental design of the generation of Ar-KO CD8 T cells in vitro using CRISPR/Cas9. Purified CD8 T cells were electroporated with Cas9/gRNA complex (NT or AR gRNA), and put in culture in vitro for 3 days in plates coated with α-CD3 and α-CD28. 3 days later, stimulated cells were harvested, and RNA was extracted or cells were restimulated in vitro for 5 h with PMA/Ionomycin, followed by ICCS (made with www.BioRender.com). c, Ar mRNA levels by qPCR in CD8 T cells electroporated with non-targeting (NT) or Ar gRNA/Cas9 after 3 days of in vitro stimulation. Data representative of 4 independent experiments with 3 replicate wells. d, Representative flow cytograms of IFNγ and TNFα expression after restimulation with PMA/Ionomycin. e, Schematic of LCMV experiment (made with www.BioRender.com), 3 animals per group. f, Ar mRNA levels in purified P14 at day 7 post adoptive transfer (from experiment described in Fig. 4e–g). Data representative of 2 independent experiments with 3 replicate wells. g, PD1 MFI and percent IFNγ+ in P14 in the blood at day 7 post adoptive transfer. Error bars represent S.D. for c and f, and S.E.M for g.
