ABSTRACT
Potent antimicrobial metabolites are produced by filamentous fungi in pure culture, but their ecological functions in nature are often unknown. Using an antibacterial Penicillium isolate and a cheese rind microbial community, we demonstrate that a fungal specialized metabolite can regulate the diversity of bacterial communities. Inactivation of the global regulator, LaeA, resulted in the loss of antibacterial activity in the Penicillium isolate. Cheese rind bacterial communities assembled with the laeA deletion strain had significantly higher bacterial abundances than the wild-type strain. RNA-sequencing and metabolite profiling demonstrated a striking reduction in the expression and production of the natural product pseurotin in the laeA deletion strain. Inactivation of a core gene in the pseurotin biosynthetic cluster restored bacterial community composition, confirming the role of pseurotins in mediating bacterial community assembly. Our discovery demonstrates how global regulators of fungal transcription can control the assembly of bacterial communities and highlights an ecological role for a widespread class of fungal specialized metabolites.
KEYWORDS: Penicillium, fungal metabolites, microbiome
INTRODUCTION
Fungal specialized (or secondary) metabolites (SMs) gained attention in the 1900s following the discovery of the world’s first antibiotic, penicillin, produced by a Penicillium isolate (1). Since then, fungal SMs have come to play pivotal roles in medicine, agriculture, and biotechnology. Fungal SMs have been well-characterized in axenic cultures, including genetic (2–5), biochemical (6), and physiological aspects of their regulation and production (7–9). In cocultures, fungal SMs have been suggested to play roles as signaling molecules that mediate the communication of the fungus with its surroundings (10–13), virulence factors to support pathogenic lifestyles (14, 15), microbial inhibitors that shape the competition with other microorganisms for finite resources (16–18), or defenses against fungivores (19, 20).
While these studies are critical first steps in understanding the potential functions of fungal SMs, the ecological roles of these compounds in multispecies communities are largely unknown. The activities of fungal SMs identified under laboratory conditions may not translate to natural communities if concentrations used in vitro are not reflective of those found in nature or if other community members are able to inactivate or alter these compounds. Purified and concentrated fungal SMs can inhibit the growth of some bacteria and fungi in large quantities in simplified lab environments (21, 22), but how naturally secreted fungal SMs operate in microbial communities is unknown. Fungal SMs could structure multispecies communities by mediating ecological interactions that favor certain species over others, resulting in a shift in community composition. We are unaware of studies that have demonstrated this scenario.
One approach for identifying fungal SMs that mediate bacterial communities is by altering the activity of global regulators of fungal metabolites. In the fungal phylum Ascomycota, the production of many SMs is under the control of the global regulatory protein of the trimeric velvet complex, LaeA (23, 24). A growing body of research has emphasized the role of LaeA in regulating SM production in monocultures of Aspergillus, Fusarium, Penicillium, and other fungal genera (25–28). LaeA also regulates many biosynthetic gene clusters (BGCs), including the aflatoxin and cyclopiazonic acid gene clusters in Aspergillus flavus (25, 29), and the patulin gene cluster in Penicillium expansum (28). Besides its impact on the metabolome, LaeA was also reported to regulate other traits in filamentous fungi, such as conidiation (30), conidial morphogenesis (31), and sclerotia formation (32). LaeA-regulated fungal traits may play important ecological roles in multispecies microbial communities, but most studies of LaeA biology have been conducted with fungal monocultures. Several studies have explored how LaeA can mediate fungal strain competition and host-microbe interactions (8, 33), but the ecological roles of LaeA in the assembly of polymicrobial communities has not been characterized.
Cheese rinds are microbial ecosystems where LaeA-regulated SMs could have significant ecological impacts. These ecosystems are composed of bacteria, yeasts, and filamentous fungi and form on the surface of many styles of cheese, including bloomy, washed, and natural rind cheeses (34). Penicillium spp. are frequently encountered in cheese rinds where they can be inoculated as industrial starter cultures (e.g., P. camemberti in Camembert or Brie) or can colonize cheese from natural populations of fungi (e.g., P. biforme, P. solitum, and P. nalgiovense in tomme style cheeses and clothbound cheddars [34, 35]). Species within this genus are prolific producers of SMs, including polyketides (e.g., patulin), nonribosomal peptides (e.g., roquefortine), and terpenes (e.g., expansolide) (36). While many Penicillium metabolites are valued as pharmaceuticals, such as the antibiotic penicillin (1) and the cholesterol-lowering drug lovastatin (37), others are considered mycotoxins, including the carcinogenic ochratoxin A (38), cyclopiazonic acid (39), and patulin (40). Penicillium species isolated from cheese rinds produce an extensive range of SMs, including mycotoxins (41, 42), and have been shown to impact the growth of neighboring bacteria (43–45), suggesting a potential of fungi to control bacterial community diversity through antibiotic production.
In the present study, we determined the ecological significance of fungal SMs in the cheese rind model system by inactivating LaeA in Penicillium sp. strain MB. The exact species identity of this fungus is currently unknown due to limited availability of whole-genome sequences of isolates from this section of the Penicillium phylogeny; a previous whole-genome sequencing analysis of this strain placed it close to Penicillium polonicum (42). When this fungus was originally isolated from a natural-rind cheese, it was the dominant filamentous fungus growing on the cheese and was preventing typical growth of the normal fungal and bacterial communities found in natural-rind cheeses. Given its negative impacts on the cheese rind, we predicted that this strain produced metabolites that could alter microbial community assembly. When we deleted laeA in this strain, an in vitro cheese rind bacterial community increased in total abundance and shifted in composition to resemble bacterial communities grown without the fungus. Both transcriptomic analysis and metabolite profiling pointed to pseurotins as putative LaeA-regulated antibacterial compounds. Inactivation of pseurotin production in the WT strain through the disruption of the gene encoding the hybrid PKS-NRPS enzyme required for pseurotin synthesis eliminated much of the antibacterial activity and caused a shift in bacterial community composition that was similar to the ΔlaeA strain. This study demonstrates the ecological relevance of LaeA-regulated fungal SMs, their roles in shaping the assembly of multispecies bacterial communities, and their possible influence on the development of human food commodities.
RESULTS
The deletion of laeA impairs several physiological traits in Penicillium sp. strain MB.
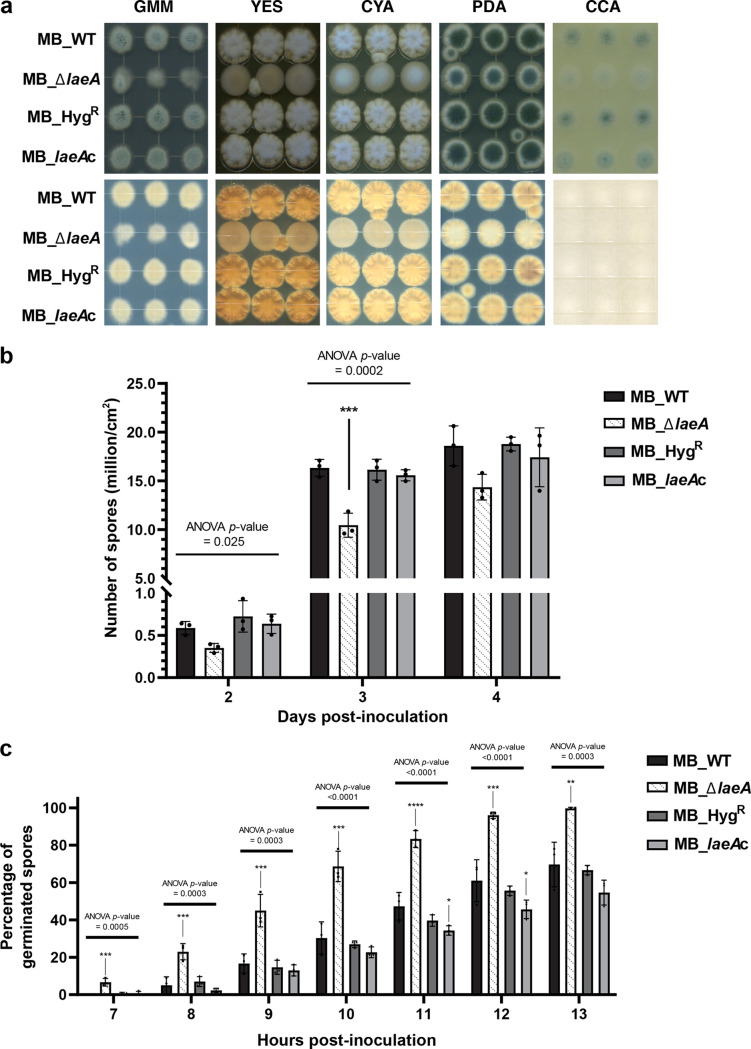
The deletion and complementation of laeA in Penicillium sp. strain MB resulted in four strains: MB_WT, MB_ΔlaeA, MB_HygR (to test for effects of inserting the hygromycin selectable marker into the genome), and MB_laeAc (complementation of the ΔlaeA strain) (see Fig. S1a to d in the supplemental material). The MB_ΔlaeA strain had altered pigmentation and reduced spore production relative to MB_WT, and complementation of the ΔlaeA strain (MB_laeAc) restored the WT phenotype (Fig. 1a and b). Differences in pigmentation were most striking on cheese curd agar (CCA), a medium that mimics cheese surface environments in cheese aging facilities (43, 46). No differences in pigmentation or sporulation were observed between the MB_WT, the MB_HygR, and the MB_laeAc strains.
FIG 1.
Deletion of laeA affects both morphological and physiological characteristics of Penicillium sp. MB strains (a) Colony aspect of Penicillium sp. MB strains grown on different media for 5 days at 25°C. GMM = glucose minimal medium, YES = yeast extract medium, CYA = Czapek Yeast Autolysate Agar, PDA = potato dextrose agar, CCA = cheese curd agar. Top and bottom views of the agar plates are shown in the top and bottom panels, respectively. (b) Spore counts for each strain over four days growth on GMM agar medium. The number of spores were assessed per standard area that was sampled by plugging the fungal colony using the shaft-attaching end of a p5000 pipette tip. (c) Percentage of germinated spores over 13 hours growth in GMM broth. Counting of germinated spores started 3 hours post-inoculation. There were no germinated spores between hours 3 to 6. In the bar graphs, the error bars represent one standard error of the mean and each dot represents a biological replicate (n = 3). One-way analysis of variance (ANOVA) was performed for each day for the sporulation data and each hour for the germination data. Dunnett’s multiple comparison test was used and compared to the MB_WT strain. (****) indicates P < 0.0001, (***) indicates P < 0.001, (**) indicates P < 0.01, (*) indicates P < 0.05, and no asterisk indicates not significant. For exact P-values for each treatment, see Data Set S1.
Genetic modifications conducted in Penicillium sp. strain MB. (a) Schematic representation of the genetic construct for laeA deletion in strain MB. The construct is constituted of a gene conferring resistance to hygromycin under TrpC promoter and terminator. (b) Southern blot analyzes of genomic DNA from the WT, the ΔlaeA, and HygR strains. The positions of the restriction enzyme cutting sites used for the Southern blot are shown on the construct schematic. Portions (10 μg) of total DNA from each strain were digested with the appropriate enzymes and subjected to Southern blot analysis using, respectively, the 5′ flank fragment (blue) and the 3′ fragment (grey) as probes. The 1-kb DNA ladder from New England Biolabs was used to determine the size of the expected bands. (c) Restriction map of plasmid pBC-phleo containing the ble gene for phleomycin resistance controlled by the Aspergillus nidulans gpdA promoter and the Saccharomyces cerevisiae CYC1 terminator. Restriction site used for cloning of the laeA gene is shown in red. (d) Southern confirmation of the single insertion of laeA using NotI as a restriction enzyme. The positive control corresponds to the digested PJT3 plasmid. (e) Map of plasmid pCS01 from which the hygromycin deletion cassette was amplified and used to knock-out the psoA gene in Penicillium sp. strain MB. (f) Scheme of the experimental strategy adopted to knockout the psoA gene in the Penicillium sp. strain MB using CRISPR-Cas9 system. The CRISPR components were delivered to the strain MB_WT protoplasts as a ribonucleoprotein (RNP) complex consisting of the Cas9 nuclease and single-guide RNA (sgRNA) targeting the PKS/NRPS conserved domain in the psoA gene sequence. The RNP complex will bind to the target sequence at the specific site and cut DNA double strands. The DNA double-strand breaks will be repaired by homology directed repair using the donor DNA template containing 1 kb of homology arms flanking the hph gene that confers resistance to hygromycin. (g) Resequencing the genomes of the ΔpsoA strains revealed the deleted regions by the CRISPR-Cas9-mediated knockout. Read mappings for the two different mutants are shown, and the deleted regions are represented by the large gap in both mappings. DNA resequencing reads are indicated by black lines. Download FIG S1, TIF file, 4.8 MB (4.8MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The deletion of laeA resulted in an accelerated germination of spores, reaching 100% germination 13 h post-inoculation, while none of the control strains were able to achieve complete spore germination (Fig. 1c). Many fungi produce self-inhibitors that reduce germination rates, especially at high inoculum levels; we speculate that the deletion of laeA may results in the diminishment of these self-inhibitors (47). The deletion of laeA also resulted in reduced growth (as measured by colony diameter) regardless of the media (see Fig. S2). Collectively, these growth and development data demonstrate that LaeA regulates fungal traits that could have consequences for neighboring microbes.
Growth rates of Penicillium spp. on YES, PDA, CYA, and CCA. (a to c) Growth on YES medium (a), growth on PDA medium (b), and growth on CYA medium (c) are represented. In the box plots, the bar represents the standard errors of the means, and each dot represents a biological replication (n = 3). One-way analysis of variance (ANOVA) was performed for each bacterial community. Dunnett’s multiple-comparison test was used, and the results were compared to the WT strain. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; no asterisk indicates not significant (ns). For the exact P values for each treatment, see Data Set 1. (d) Growth on CCA, where points and connecting lines with standard errors of the means bars were used from two independent experiments with five biological replications each. In experiment 2, replication 5 was removed due to no growth of the bacteria in the control. Two-way ANOVA was performed for each fungal strain. Dunnett’s multiple-comparison test was used and compared to the WT strain. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; no asterisk indicates not significant (ns). For the exact P values for each treatment, see Data Set 1. Download FIG S2, TIF file, 0.9 MB (886.2KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Penicillium traits regulated by LaeA mediate cheese rind bacterial community assembly and microbial interactions.
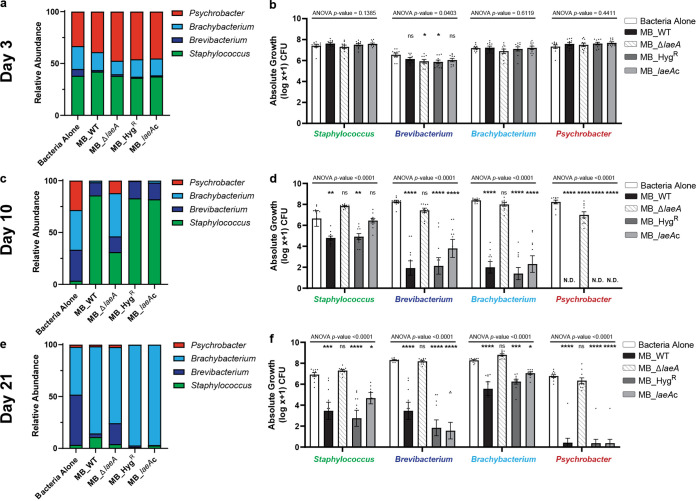
To test how LaeA’s regulation of physiological or metabolic traits alters the development of cheese rind microbiomes, we grew all four Penicillium strains (MB_WT, MB_ΔlaeA, MB_HygR, and MB_laeAc) with a four-member bacterial community that represents the dominant taxa found in typical natural rind cheeses. We compared total bacterial community abundance (as total CFU) and bacterial community composition (relative abundance of each community member) at 3, 10, and 21 days post-inoculation across five different treatments: (i) bacteria alone, (ii) bacteria + Penicillium WT strain, (iii) bacteria + Penicillium ΔlaeA strain (MB_ΔlaeA), (iv) bacteria + Penicillium hygromycin resistance control strain (MB_HygR), and (v) bacteria + Penicillium laeA complement strain (MB_laeAc).
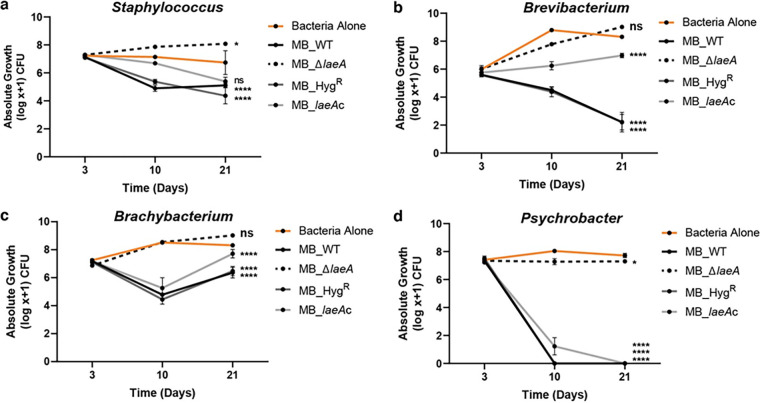
After 3 days of incubation, community composition was similar across all treatments (Fig. 2a and b). However, the presence of MB_WT, MB_HygR, and MB_laeAc strains caused a significant restructuring of the bacterial community through their strong inhibitory effects at day 10 post-inoculation (Fig. 2c; see the Fig. 2 legend for PERMANOVA [permutational multivariate analysis of variance] statistics). The bacteria alone treatments were dominated by Actinobacteria (Brevibacterium [30%] and Brachybacterium [38%]), whereas the MB_WT, MB_HygR, and MB_laeAc treatments were Staphylococcus-dominated (>82% of the population) (Fig. 2c). In the presence of MB_WT, the absolute growth for all bacteria was significantly reduced, with Brevibacterium and Brachybacterium having nearly a 4-fold decrease in absolute growth, and Psychrobacter populations not reaching detectable levels (Fig. 2d). Comparable results were observed with the Penicillium MB_HygR and MB_laeAc strains. Strikingly, the abundance and structure of the bacterial community in the presence of MB_ΔlaeA was similar to the community grown in the absence of the fungus (a range of 57 to 68% Actinobacteria) (Fig. 2c). The shifts in community composition and dominance of Actinobacteria in the bacterium-alone and MB_ΔlaeA communities persisted through day 21 (Fig. 2e and f).
FIG 2.
Inactivation of LaeA increases bacterial diversity and abundance in cheese rind communities. The stacked barcharts show the shifts in the relative abundances of a typical rind bacterial community in the presence of four strains of Penicillium sp. strain MB after (a) 3, (c) 10, and (e) 21 days of incubation. Data are mean relative abundance from two independent experiments with five biological replicates each. The compositions of the Bacteria Alone and MB_ΔlaeA communities were not significantly different from one another, but were different from MB_WT, MB_HygR, MB_laeAc at 10 and 21 days (Day 3 PERMANOVA F = 0.9958, p = 0.425; Day 10 PERMANOVA F = 23.12, P < 0.0001; Day 21 PERMANOVA F = 11.14, P < 0.0001). The bar graphs show absolute abundances of individual bacterial community members from the same experiments as in (a), (c), and (e) in the presence of Penicillium sp. strain WT and mutants (MB_ΔlaeA, MB_HygR, and MB_laeAc) after (b) 3 (d), 10 and (f) 21 days of incubation. Each bar represents the mean with standard errors and each dot represents a biological replicate. One-way analysis of variance (ANOVA) was performed for each bacterium. Dunnett’s multiple comparison test was used and compared to the WT strain. (****) indicates P < 0.0001, (***) indicates P < 0.001, (**) indicates P < 0.01, (*) indicates P < 0.05, and no asterisk indicates not significant (ns). For exact P-values for each treatment, see Data Set S1.
Penicillium species often co-occur with yeasts in cheese rinds (34, 43), and the presence of another fungus may dampen the inhibitory effects of Penicillium sp. strain MB on the bacterial communities. To test this, we repeated all community experiments with the addition of the common cheese rind yeast, Debaryomyces hansenii. We observed nearly identical patterns of inhibition of bacteria by MB_WT and loss of inhibition in MB_ΔlaeA communities in the presence of D. hansenii (see Fig. S3). This demonstrates that even with a more realistic two species fungal community, laeA-regulated traits of Penicillium sp. MB can control bacterial community diversity.
Community analysis in the presence of another common cheese rind fungal community member, the yeast Debaryomyces hansenii. Histograms show the shifts in a typical rind community (Staphylococcus, Brevivacterium, Brachybacterium, and Psychrobacter) in the presence Penicillium sp. strain MB_WT and mutants (ΔlaeA, HygR, and laeAc) and D. hansenii after 3 (a), 10 (c), and 21 (e) days of incubation. The data are mean relative abundances from two independent experiments. Histograms show the inhibition of community members grown in the presence Penicillium sp. strain MB_WT, mutant strains, and D. hansenii after 3 (b), 10 (d), and 21 (f) days of incubation. The data are from two independent experiments, with five biological replications each. In the box plots, the bar represents the standard errors of the means, and each dot represents a biological replication. One-way ANOVA was performed for each bacterial community. Dunnett’s multiple-comparison test was used and compared to the WT. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; no asterisk indicates not significant (ns). For the exact P values for each treatment, see Data Set 1. Download FIG S3, TIF file, 1.8 MB (1.8MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
To determine the role of LaeA in mediating pairwise interactions between Penicillium sp. MB and each of the individual bacterial species, we cocultured each bacterium separately with the four Penicillium sp. MB strains on cheese curd agar. After 21 days, strains that had a functional LaeA (MB_WT, HygR, and MB_laeAc) inhibited the growth of all four bacteria (Fig. 3) with an inhibition hierarchy (log10% decrease alone versus MB_WT) of Psychrobacter (100% inhibition) > Brevibacterium (73% inhibition) > Staphylococcus (24% inhibition) > Brachybacterium (21% inhibition). These data demonstrate that Penicillium sp. MB strongly and directly inhibits the growth of individual cheese rind bacteria, and this inhibitory effect is mediated by LaeA.
FIG 3.
Pairwise interaction assays showing inhibition of bacterial strains grown individually in the presence of each of the four strains of Penicillium sp. strain MB. Data for (a) Staphylococcus, (b) Brevibacterium, (c) Brachybacterium, and (d) Psychrobacter are shown. Points indicate means and error bars indicate standard errors from two independent experiments with five biological replicates each. In experiment 2, replication 5 was removed due to no growth of the bacteria in the control. Two-way analysis of variance (ANOVA) was performed for each bacterial community. Dunnett’s multiple comparison test was used and compared to the WT strain. (****) indicates P < 0.0001, (***) indicates P < 0.001, (**) indicates P < 0.01, (*) indicates P < 0.05, and no asterisk indicates not significant (ns). For exact P-values for each treatment, see Data Set S1.
Collectively, these community and pairwise data demonstrate that LaeA regulates some aspect of Penicillium sp. strain MB physiology or metabolism that results in differential inhibition of bacterial species growth in the cheese rind community. The LaeA-regulated factor(s) have the ability to completely transform the composition of the bacterial community.
RNA sequencing reveals global alterations in the expression of specialized metabolite genes in the ΔlaeA strain.
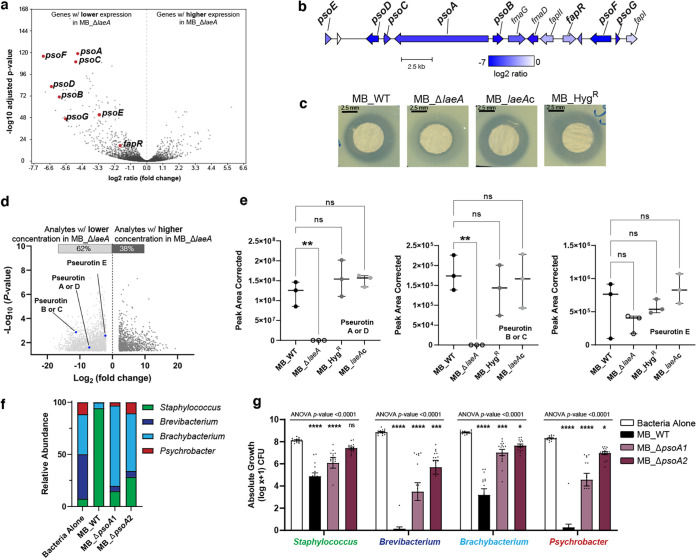
To identify SM biosynthetic gene clusters (BGCs) and other genes regulated by laeA in Penicillium sp. strain MB that might be driving bacterial-fungal interaction outcomes, we performed gene expression profiling using RNA sequencing (RNA-seq). RNA-seq libraries prepared from mycelium of MB_WT and MB_ΔlaeA harvested 48 h post-inoculation from CCA were examined for differential gene expression. This time point was selected because physiological differences between both strains were visually apparent (differences in pigmentation and spore production), and significant amounts of high-quality RNA could be obtained. Using a log2 ratio cutoff of 2 (corrected P value of <0.05), we identified 253 genes with decreased expression and 57 genes with increased expression in the MB_ΔlaeA strain (Fig. 4a; see also Data Set S2a). No transcripts for laeA were detected in the MB_ΔlaeA strain confirming the successful deletion of this gene.
FIG 4.
Deletion of laeA impairs production of metabolites with associated antimicrobial properties. (a) RNA-sequencing results showed a significant downregulation of biosynthetic genes in the pseurotin putative gene cluster when laeA was deleted. (b) Organization of the putative pseurotin biosynthetic gene cluster in Penicillium sp. strain MB and log2 ratio of expression in MB_ΔlaeA. Gene names in bold are those known to be involved in pseurotin production in Aspergillus species. (c) Visualization of zones of inhibition of crude extracts collected from all four Penicillium isolates on YES medium against Brachybacterium alimentarium evaluated by the disk diffusion method. See Fig. S4 for additional zone of inhibition data for other bacterial species. (d) Volcano plots representing the number of analytes significantly regulated in MB_ΔlaeA compared to the MB_WT strain on YES medium. The blue dots indicate pseurotin A and the other putative pseurotins identified in this analysis. (e) Comparison of the peak area corrected values for pseurotin A and the other putative pseurotins between strains. Each bar represents the means with +/− one standard error, and each dot represents a biological replicate (n = 3). One-way analysis of variance (ANOVA) was performed for each Penicillium strain. Dunnett’s multiple comparison test was used and compared to the MB_WT strain. (**) indicates P < 0.01 and no asterisk indicates not significant (ns). (f) Inactivation of psoA led to loss of antibacterial activity and restored bacterial community composition. The compositions of MB_ΔpsoA-1 and MB_ΔpsoA-2 bacterial communities were not significantly different from one another but were different from Bacteria Alone and MB_WT (PERMANOVA F = 49.97, P < 0.0001). (g) Histograms showing the inhibition of community members grown in the presence of the four strains of Penicillium sp. strain MB at 10 days post-inoculation. Data are mean relative abundance from three independent experiments with five biological replications each. In the box plots, the bar represents the standard errors of the means and each dot represents a biological replication. One-way analysis of variance (ANOVA) was performed for each bacterial community. Dunnett’s multiple comparison test was used and compared to the WT strain. (****) indicates P < 0.0001, (***) indicates P < 0.001, (**) indicates P < 0.01, (*) indicates P < 0.05, and no asterisk indicates not significant (ns). For exact P-values for each treatment, see Data Set S1.
Using an enrichment analysis of differentially expressed genes’ GO terms, we identified depsipeptide, emericellamide, lactone, aspartic peptidase, indole alkaloid, fumagillin, epoxide, and alkaloid biosynthesis as pathways with greater than 30% of genes in a pathway being significantly downregulated (see Data Set S2b). All these biosynthetic pathways play major roles in SM production of fungi. The GO terms glucose, hexose, monosaccharide, and nucleobase transporters were enriched in the upregulated genes of the MB_ΔlaeA strain (see Data Set 2b).
We used the list of downregulated genes and antiSMASH predictions to identify specific BGCs that might be related to the observed inhibition of bacterial growth. BGCs were identified by locating groups of adjacent downregulated genes with annotations for hallmarks of biosynthetic gene clusters, including polyketide synthases, nonribosomal peptide synthetases, terpene cyclases, and prenyltransferases (48). The most downregulated BGC in the MB_ΔlaeA strain contained genes homologous to the A. fumigatus fumagillin/pseurotin supercluster (49) (Fig. 4a). Pseurotins are a family of fungal alkaloids that have been reported as having antibacterial and insecticidal activities (50). A detailed analysis of the pseurotin BGC describing its genetic organization was reported in A. fumigatus (49), but pseurotin biosynthetic pathways and modes of antibacterial action are not fully known due to the presence of many intermediate compounds (51). The Penicillium sp. MB genome contains a 16-gene cluster with a complete set of predicted genes for pseurotin biosynthesis, including psoA, psoB, psoC, psoD, psoE, psoF, and psoG (Fig. 4b). Some genes essential for fumagillin biosynthesis, including the terpene cyclase gene (fmaA) are missing, suggesting the strain could synthesize pseurotin but not fumagillin (Fig. 4b). All these genes were highly downregulated (−4.7 to −7 log2-fold) and were among the most downregulated genes in the MB_ΔlaeA strain (Fig. 4a). Based on antiSMASH predictions (see Data Set S2c), the Penicillium sp. strain MB genome contains 42 regions that could encode putative specialized metabolites. Some of these other BGCs that were downregulated in the MB_ΔlaeA strain included a putative aspterric acid and quinolone BGC not known to have antibacterial activity, as well as other BGCs encoding putative metabolites (see Data Set S2a). None of these BGCs were downregulated to the same extent as the pseurotin BGC.
Loss of laeA alters the production of all members of the 1-oxa-7-aza-spiro[4,4]non-2-ene-4,6-dione class of antibacterial natural products.
Our findings highlighted a significant role for laeA in the assembly of microbial communities that may be partly due to a fitness cost associated with physiological traits of this mutant (Fig. 1). The downregulation of several BGCs in the MB_ΔlaeA strain suggested that differences in community structure between treatments could be due to antibacterial activities of laeA-regulated metabolites. To explore this hypothesis, we assessed metabolomic changes resulting from laeA deletion after 14 days of growth on four different media (CYA, PDA, YES, and CCA). Crude extracts collected from all four Penicillium strains were later screened for antibacterial activity using the disk diffusion method against the same bacterial species used in the community and pairwise interaction assays.
Crude extracts collected from MB_WT growing on YES medium showed the highest antibacterial effects, with a clear circular zone of inhibition on all tested bacteria (Fig. 3c; see also Fig. S4a). Interestingly, crude extracts from MB_ΔlaeA cultures generated significantly smaller zones of inhibition than extracts from control strains regardless of the bacterial strain tested (see Fig. S4a). Crude extracts collected from cultures of the MB_ΔlaeA and the control strains did not show significant differences in their ability to inhibit bacterial growth, except against Psychrobacter (see Fig. S4b to d). Extracts collected from CCA cultures showed the lowest inhibitory effects on all tested bacteria, likely due to the high amount of fat in this medium resulting in low recovery of metabolites from the organic phase (see Fig. S4d). Similar problems with metabolite recovery have previously been attributed to the high fat content in milk (52). Therefore, we focused on exploring the metabolomic changes on YES medium to pinpoint the laeA-regulated metabolite(s) responsible for the shift in bacterial community composition.
Antibacterial activities of filtered crude extracts from cultures on different media. Crude extracts were collected from cultures of the different Penicillium strains on YES (a), PDA (b), CYA (c), and CCA (d) media. The diameter of the zone of inhibition was measured and analyzed. In the box plots, the bar represents the standard errors of the means, and each dot represents a biological replication (n = 3). One-way ANOVA was performed for each bacterial community. Dunnett’s multiple-comparison test was used and compared to the WT strain. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; no asterisk indicates not significant (ns). For the exact P values for each treatment, see Data Set 1. Download FIG S4, TIF file, 0.9 MB (916.3KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Analysis of metabolomic data on YES medium showed that the deletion of laeA led to a 2-fold decrease in the production of 62% of the 2,703 significantly differentially produced analytes and 2-fold increase in the production of 38% of all significantly differentially produced analytes in negative ionization mode (Fig. 4d). Pseurotins have a 1-oxa-7-aza-spiro[4,4]non-2-ene-4,6-dione core and multiple forms exist (A through F) with slight modifications to this core (53). Using a list of chemical formulas assigned to known metabolites produced by this Penicillium species and closely related species, we were able to identify peaks that correspond to putative pseurotins. Strikingly, the antibacterial metabolite pseurotin A showed a 140-fold decrease in the ΔlaeA mutant compared to the WT strain (P = 0.025) (Fig. 4e). The identification of pseurotin A (C22H25NO8) was confirmed by high-resolution mass spectrometry (HRMS) [M-H]– (m/z 430.1503, calculated for C22H24NO8 430.1507) and by HRMS/MS fragmentation. The mass spectrum showed the two fragmentation ions (m/z 270.0768 and 308.1137) consistent with the mass spectrum ion previously reported for pseurotin A (49). We could not perform experimental confirmation of other putative pseurotins due to the lack of commercially available standards or fragmentation databases. The published chemical formulas of pseurotins B, C, D, and E (54) were applied to MAVEN, which identified a significant reduction of production of these putative metabolites in the absence of laeA (Fig. 4e).
The significant decrease in the synthesis of pseurotin A and the other pathway derivatives on YES medium matched the transcriptomic data showing a downregulation of the pseurotin gene cluster in the MB_ΔlaeA strain (Fig. 4a). Combined with previous reports of antibacterial properties of pseurotin (50), our concurring datasets suggested that pseurotins could explain the striking antibacterial activity of this Penicillium isolate.
Inactivation of pseurotin production leads to a loss of antibacterial activity and restores bacterial community composition.
To test whether pseurotins were the fungal metabolites regulating bacterial community structure, we disrupted the hybrid PKS-NRPS synthase gene psoA required for pseurotin production in Penicillium sp. strain MB using a CRISPR-Cas9 system (see Fig. S1e and f). The disruption of this gene in two independent mutants (MB_ΔpsoA-1 and MB_ΔpsoA-2) and the absence of Cas9 off-target cleavage events were confirmed using whole-genome resequencing (see Fig. S1g). Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of MB_ΔpsoA-1 and MB_ΔpsoA-2 confirmed a lack of pseurotin A production (see Fig. S5). In contrast, MB_WT produced on average 1.36 mg of pseurotin A (see Data Set S1).
LC-MS data showing the absence of pseurotin production in the crude extracts of both MB_ΔpsoA strains compared to the MB_WT strain. (a) Chromatograms showing the absence of pseurotin A and putative pseurotin D peaks in the ΔpsoA mutants. (b) Fragmentation mass spectrum in positive ionization mode of pseurotin A in MB_WT strain compared to pseurotin A standard. (c) Fragmentation mass spectrum in positive ionization mode putative pseurotin D in MB_WT strain. The MS/MS spectrum shown for the putative pseurotin D is for reference purposes only and represents the differentially eluting isomer that was also affected by the genetic knockouts performed here. Download FIG S5, TIF file, 0.8 MB (859.3KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
When we repeated the bacterial community assembly assays described above with the ΔpsoA mutants, we saw two striking patterns (Fig. 4f and g). First, the two mutants did not reduce total bacterial growth as much as the MB_WT strain; while MB_WT had a 34.7% reduction in total CFU across all four bacterial species, the average total bacterial reduction across the two pseurotin mutants was only 13.9% (Fig. 4g). As we observed earlier (Fig. 2), Brevibacterium, Brachybacterium, and Psychrobacter were most inhibited by the MB_WT strain. Similar to coculture with MB_ΔlaeA, the ΔpsoA mutants had minor inhibitory impacts on these bacterial genera (Fig. 4g). Second, we observed that bacterial community composition was significantly different between the MB_WT and the MB_ΔpsoA-1 and MB_ΔpsoA-2 mutant communities. As with the ΔlaeA knockout above, the bacterial community composition shifted from dominance of Staphylococcus in the MB_WT community to a mix of all bacterial species in the MB_ΔpsoA-1 and MB_ΔpsoA-2 communities (Fig. 4f). These data combined with our transcriptomic and metabolomic data above demonstrate that the pseurotin BGC is regulated by LaeA in this Penicillium species and that the loss of pseurotin production eliminates much of the strong inhibitory effect of this fungus on bacterial community assembly.
Components of the pseurotin biosynthetic gene cluster are widely distributed across the Ascomycota.
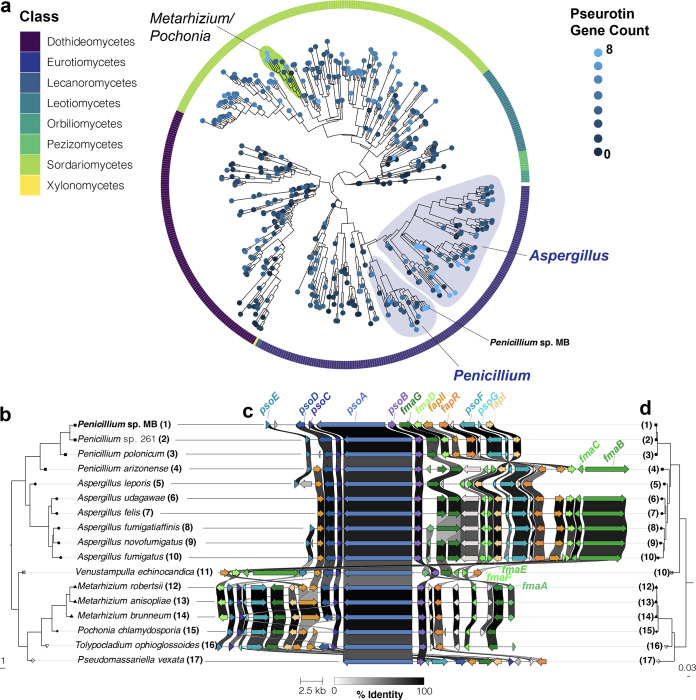
Considering the strong ecological impacts of the pseurotin BGC, we used comparative genomics to identify other fungi whose genomes encode this cluster or closely related clusters with similar antimicrobial properties. Previous studies have demonstrated that the pseurotin/fumagillin supercluster was subject to rearrangements and gene loss in A. clavatus, Neosartorya fischeri, and Metarhizium anisopliae (49). Scanning across a broad sample of fungal genomes, we found that pseurotin genes are only present in the Ascomycota (Fig. 5; see also Fig. S6). Only a small subset of fungi are predicted to have all eight pseurotin genes, including species in the genera Aspergillus, Penicillium, Metarhizium, Tolypocladium, and Venustampulla (see Data Set S3). Of the 77 species of Aspergillus in this data set, only 11 have all 8 genes. Of the 24 species of Penicillium, only 2 (in addition to Penicillium sp. strain MB) have all 8 genes (Fig. 5a). Another closely related Penicillium strain that we isolated from a separate cheese production facility in the United States (Penicillium sp. strain 261) also has the full pseurotin BGC (Fig. 5b).
FIG 5.
Components of the pseurotin biosynthetic gene cluster are widespread across Ascomycota, but the full cluster is mainly found in Aspergillus and Penicillium species. (a) The subclade of Ascomycota identified as having higher prevalence of pseurotin cluster genes (Fig. S6) presented with a single leaf per species. Phylogenetic relationships were determined from the consensus of 290 maximum-likelihood trees constructed for benchmarking single copy orthologs (BUSCOs). The outer ring indicates the taxonomic order that species pertain to. Terminal branch lengths are not calculated. The number of pseurotin genes was determined using reciprocal best-hit BLAST analysis using the Aspergillus fumigatus pseurotin genes psoA, psoB, psoC, psoD, psoE, psoF, psoG, and fapR is indicated as the intensity of the tip color. (b) A phylogeny constructed in the same way as (a) representing a subset of species identified as having seven or eight pseurotin genes that were manually selected to represent taxonomic breadth. (c) An alignment of pseurotin gene clusters. Genes in the pseurotin cluster are labeled in blue while constituents of the intertwined fumagillin cluster are indicated in green. Regulatory genes of these clusters are labeled in orange. Coloration of genes is based on homology searches and visualizations performed by clinker. The weight of lines between homologous genes indicates the percent identity; genes sharing identity below 30% are not indicated. (d) A maximum likelihood phylogeny constructed from the concatenation of PsoA, PsoB, and PsoC sequences. Note that phylogenetic relationships between (d) and (b) are compatible, suggesting vertical transmission of the pseurotin gene cluster. The strain information of the species used in this analysis are provided in Data Set S3.
Frequency of genes associated with the pseurotin gene cluster (left) across fungal phyla (right). A total of 1.463 fungal genomes representing 808 species were obtained from the NCBI. Phylogenetic relationships between genomes were determined from the consensus of 290 maximum-likelihood trees constructed for single-copy orthologs (BUSCOs). Terminal branch lengths were not calculated. The number of pseurotin genes present in each genome was determined using reciprocal best-hit BLAST analysis using the Aspergillus fumigatus pseurotin genes psoA, psoB, psoC, psoD, psoE, psoF, psoG, and fapR. The orange arrow indicates a monophyletic clade within Ascomycota that was visually identified as having a higher prevalence of pseurotin genes. Download FIG S6, TIF file, 0.9 MB (873.6KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Given the taxonomic breadth of fungi containing the pseurotin cluster, we sought to understand the evolutionary origins of this cluster within and outside the Ascomycota, including whether horizontal or vertical transmission explains patterns of pseurotin gene distribution. The overall history of this BGC appears consistent with vertical transmission and gene loss in some lineages; the topology of a phylogeny constructed from 290 benchmarking single copy orthologs (BUSCOs) was compatible with another phylogeny constructed using PsoA, PsoB, and PsoC sequences (Fig. 5b to d). We cannot entirely rule out the possibility of horizontal gene transfer, but such transfers would have to be ancient to allow for BGC and species histories to align. Patterns of vertical transmission are also apparent in the synteny of the BGC. For example, an inversion of several genes at one end of the cluster (fmaD-fapI) that is present in most Penicillium spp. but not in Aspergillus spp. is missing in the early-divergent Penicillium arizonense (Fig. 5c), suggesting that this mutation occurred sometime after the diversification of this genus.
To explore the possibility of an ancient horizontal gene transfer event from outside the fungi, we BLASTed the PsoA protein against all NCBI databases, excluding fungi. The highest-scoring hit from this search (94% of the query cover, 32.38% identity) was an uncharacterized hypothetical protein that is annotated as PKS in the plant Carpinus fangiana (accession KAB8336832.1). However, the reciprocal BLAST of this protein against fungi found better hits (100% coverage and ~60% identity) against other uncharacterized PKSes and putative lovastatin nonaketide synthases. While our results demonstrate that the pseurotin BGC is widespread in a subclade of Ascomycota, further analyses exploring the evolution of this gene cluster are needed.
DISCUSSION
The chemistry, genetics, and physiology of fungal specialized metabolism are widely studied and recognized, but their ecological consequences in microbiomes are largely unknown. Previous genetic and -omics approaches identified key metabolites and pathways regulated by LaeA (28, 29, 55–57), but these data have not been integrated into a comprehensive model for studying microbial interactions that influence community assembly. By toggling LaeA between on and off, we were able to identify one class of metabolites produced by a strongly inhibitory fungus that allows it to dramatically remodel the bacterial communities of cheese. Our integration of physiological assays, transcriptomics, and metabolomics more broadly demonstrates how LaeA can be used to identify fungal metabolites that control the assembly of microbiomes.
The intertwined pseurotin and fumagillin supercluster was initially identified in Aspergillus fumigatus and reported to be under the control of LaeA (49). Our study shows that this BGC exists in the cheese-isolated Penicillium sp. strain MB and is also positively regulated by LaeA (Fig. 4a and b). This metabolite was previously found to exhibit a range of bioactivities at moderate to high concentrations (up to 50 μg/mL) with potential therapeutic applications due to its immunosuppressive (58), antibacterial (50, 59), nematocidal (60), insecticidal (61), antiparasitic, and anticancer activities (62). Previous studies of antibacterial properties have focused on purified versions of the compound and have not used natural production from a host for assays. In our analysis, the production of pseurotin A by the MB_WT strain was roughly 2.7 times higher than the concentration used in the aforementioned studies (see Data Set S1) and may explain why this fungus has such strong inhibitory effects on bacterial communities. Characterization of how other fungi that produce pseurotins impact bacterial community assembly will further clarify how this class of metabolites plays roles in multispecies microbiomes. In addition, why pseurotins have antibacterial activity is not currently known, and additional work characterizing the mode of action is needed.
In addition to its regulation and role in the cheese Penicillium isolate, we have found that components of the pseurotin BGC can be found intermittently in the Ascomycota, but not in any other fungal phyla. Pseurotin A has been reported in several different filamentous fungi, including species of Aspergillus, Penicillium, and more recently the distantly related taxon Metarhizium (51, 61, 63, 64). Our finding of all eight genes of this BGC in several other genera (Tolypocladium and Venustampulla) suggests that fungal pseurotins may be widely distributed across a range of fungal niches, from food production and indoor environments to forest soils. Additional studies of these fungi and neighboring bacteria are needed to fully characterize the ecological significance of pseurotins across ecosystems.
Our data strongly suggest that pseurotins are key LaeA-regulated metabolites that can mediate bacterial community assembly, but we acknowledge that other fungal metabolites and traits could be playing roles in this system. Other BGCs, including some putative clusters with unknown functions, were downregulated in the transcriptome of the MB_ΔlaeA strain (see Data Set S2), suggesting that they are regulated by LaeA. These may also contribute to the antibacterial effects of Penicillium sp. strain MB. Future work using untargeted metabolomics and other approaches may identify these additional LaeA-regulated metabolites in this and other filamentous fungi that affect bacterial community assembly.
Every time a consumer eats a piece of a naturally aged cheese, they ingest the many metabolites secreted into the cheese by rind microbes. Surprisingly little is known about the diversity and functions of these microbial metabolites (41, 65–68). Our work demonstrates that fungal SMs produced in cheese environments can serve as mediators of microbiome formation at concentrations relevant to naturally aged cheeses. The interactions described here occurred in a highly controlled laboratory environment and their relevance in full-scale cheese production remains to be determined. The Penicillium sp. strain MB used in this study was causing cheese production problems by inhibiting normal rind formation, suggesting that the disrupted community assembly observed in the lab could also happen in cheese caves. Many other filamentous fungi found in cheese rinds possess uncharacterized metabolites that might be responsible for similar negative outcomes. Systematic exploration of LaeA-regulated metabolites and their ecological roles will not only discover microbial mechanisms underlying traditional cheese-making but will also illuminate how fungal SMs mediate microbiome composition in all environmental niches where fungi reside.
MATERIALS AND METHODS
Microbial strain isolation and culture conditions.
The Penicillium sp. strain MB used in this study was isolated from a natural rind cheese made in the United States. Its genome has been deposited in NCBI with accession number GCA_008931935.1. The exact species identification of this strain is unknown at this time because genomes of type strains of closely related species are not currently available. However, previous comparative genomic analysis suggests it is near P. polonicum in section Fasiculata of the genus Penicillium (69). The beta-tubulin marker gene (benA) of Penicillium sp. strain MB has a high similarity with two Penicillium species: 98.6% pairwise identity with a reference strain of P. cyclopium (strain CBS 14445) and 96.1% pairwise identity with a reference strain of P. polonicum (strain CBS 22228). Preliminary observations demonstrated that this Penicillium strain had potent antibacterial activity and was therefore an interesting strain to explore microbial interactions and secondary metabolite regulation through the inactivation of the global regulator of fungal secondary metabolism, LaeA.
To prepare a fresh spore suspension prior to experiments, the Penicillium sp. strain MB was activated on glucose minimal media (GMM) (70) for 7 days at 25°C, and spores were harvested in 0.01% Tween 80 and counted using a hemocytometer. Bacterial strains were also maintained as glycerol stocks and streaked on brain heart infusion agar prior to experiments. The CYA, YES, PDA, and GMM agars were prepared as previously described (70), and the CCA medium was also prepared as previously described (43).
Four bacterial strains (Staphylococcus equorum strain BC9, Brevibacterium aurantiacum strain JB5, Brachybacterium alimentarium strain JB7, and Psychrobacter sp. strain JB193) were used in bacterial community experiments. These species span three bacterial phyla (Firmicutes, Actinobacteria, and Proteobacteria) (43) that are most abundant in cheese rinds. They have been used as a model community in previous work from our lab (44, 45, 71) and have been demonstrated to have various responses to the presence of Penicillium (43, 45). This model community also has a well-defined community succession with Staphylococcus and Psychrobacter dominating early in succession (days 0 to 10) and the Actinobacteria Brevibacterium and Brachybacterium dominating later in succession (days 10 to 21). We did not use bacteria isolated from the same cheese where the Penicillium sp. strain MB was originally isolated because we were interested in identifying the basic ecological impacts of LaeA on bacterial communities, not the specific way in which this mold was interacting with bacteria on the cheese surface. By using previously well-characterized responding bacteria, we can compare our community assembly assays and interactions in this work to many previous experiments. We acknowledge that the bacteria that co-occurred with the Penicillium sp. strain MB may have had a different response compared to our model bacterial community.
Construction of gene deletion cassettes.
To knockout laeA in the Penicillium sp. strain MB, the isolate was first subjected to antimicrobial susceptibility testing toward hygromycin and phleomycin, two antibiotics commonly used by our group as selectable markers in fungal transformations. The isolate showed a confirmed sensitivity to both antibiotics. A three round PCR deletion strategy was used to replace the laeA open reading frame (ORF) in the Penicillium sp. strain MB with the hph gene, whose expression confers selection on hygromycin. The schematic representation of the laeA gene replacement with the hph gene is depicted in Fig. S1a. Each deletion cassette (5′flank-hph-3′flank) was constructed using three sequential PCRs. In the first PCR round, about 1 kb of genomic sequence that flanks either the 5′ or the 3′ end of the laeA ORF was amplified from strain MB using, respectively, the primer set PMB_KOlaeA_5′ or 3′F/R. The hph gene was amplified from plasmid pUCH2-8 using primers hph_F and hph_R. A second PCR was performed to assemble by homologous recombination the three individual fragments from the first-round PCR. The deletion cassettes were finally amplified using nested primer sets (PMB_KOlaeA_NestedF/R).
To test whether pseurotin was involved in the antimicrobial activity and shifts in bacterial community composition observed with this Penicillium strain, we knocked out the psoA gene that encodes for the hybrid polyketide synthase-nonribosomal peptide synthetase (PKS/NRPS) of the putative pseurotin biosynthetic gene cluster to disrupt the production and accumulation of all pseurotin. The same strategy described earlier was adopted for the construction of the psoA deletion cassette. The primer sets PMB_KOpsoA_5′/R and 3′F/R were used to amplify the 1-kb homology arms flanking the psoA gene on the 5′ and 3′ ends, respectively. The hph gene under the control of the Tef1 promoter and terminator was amplified from the plasmid pCS01 using the primer set Hyg-tef1F/R. The plasmid map is given and annotated in Fig. S1e. A second PCR was performed to assemble by homologous recombination the three individual fragments from the first round PCR. The deletion cassette was finally amplified using the nested primer set (PMB_psoA_nestedF/R). The sequences of the primer sets used for the construction of the deletion cassettes are shown in Table S1.
Primers used in this study for the construction of deletion cassettes and molecular cloning. Download Table S1, DOCX file, 0.02 MB (16.1KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
PEG-mediated protoplast transformation.
To generate the deletion strains, a protoplast-mediated transformation protocol routinely used by our group was employed and optimized to achieve a successful protoplasting of Penicillium sp. strain MB. Briefly, 109 fresh spores from each strain are cultured in 500 mL of GMM broth supplemented with 1 g/L yeast extract for 12 h under 25°C and 280 rpm. Newly born hyphae were harvested by centrifugation at 8,000 rpm for 15 min and hydrolyzed in a mixture of 30 mg of lysing enzyme from Trichoderma harzianum (Sigma-Aldrich) and 20 mg of Yatalase (Fisher Scientific) in 10 mL of osmotic medium (1.2 M MgSO4 and 10 mM sodium phosphate buffer). The quality of protoplast was monitored under the microscope after 4 h of shaking at 28°C and 80 rpm. The protoplast mixture was later overlaid gently with 10 mL of chilled trapping buffer (0.6 M sorbitol and 0.1 M Tris-HCl [pH 7.0]) and centrifuged for 15 min under 4°C and 5,000 rpm. Protoplasts were collected from the interface, overlaid with an equal volume of chilled STC (1 M sorbitol, 10 mM Tris-HCl, and 10 mM CaCl2) and decanted by centrifugation at 6,000 rpm for 10 min. The protoplast pellet was resuspended in 500 μL of STC and used for transformation. For protoplast transfection, 100 μL of freshly isolated protoplasts and 5 μg of linear DNA containing the deletion cassette were mixed to a final volume of 200 μL of STC buffer. The contents were mixed by gently inverting the tubes. After 50 min incubation on ice, 1.2 mL of 60% (wt/vol) PEG solution (60 g of PEG 3350, 50 mM CaCl2, and 50 mM Tris-HCl [pH 7.5]) was added to the mixture, followed by incubation for an additional 20 min at room temperature. The mixture was supplemented with 5 mL of STC and mixed into 50 mL of SMM top agar (GMM supplemented with 1.2 M sorbitol) containing hygromycin at a final concentration of 150 μg/mL. The mixture was inverted several times, and each 5-mL portion was poured onto a selective SMM bottom agar plate. The transformation plates were incubated at 25°C for 5 to 7 days.
(i) CRISPR/Cas9-mediated knockout of psoA gene.
The large size of the psoA gene (~12 kb) made the standard transformation method described for laeA deletion ineffective. Therefore, we switched to a ribonucleoprotein-CRISPR-Cas9 (RNP-CRISPR-Cas9) system. The schematic representation of the CRISPR cas9 system used for engineering the psoA knockout strain is depicted in Fig. S1f. The fungal transformation steps followed were the same as described above, the only difference being the codelivery of the Cas9-gRNA RNP complex along with the linear donor DNA (psoA deletion cassette) to the protoplasts. The CRISPR RNA (crRNA) was designed on the PKS-NRPS conserved domain in the psoA sequence using the CRISPOR webtool (http://crispor.tefor.net/) that offers off-target and efficiency predictions. The selected crRNA (5′-GGAUCGAUCUUGAACAGCAG-3′) showed a predicted targeting efficiency of 100% and 0 predicted off targets. This crRNA was subjected to an additional confirmation using the RNAfold web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) that allows to determine the gRNA secondary structure. The interpretation of the results and validation of the gRNA were done following the guidance by Hassan et al. (72). The crRNA and tracrRNA (IDT, catalog no. 1072534) were synthesized using the Alt-R CRISPR-Cas9 system from Integrated DNA Technologies (IDT; San Diego, CA). The crRNA and tracrRNA were mixed at a concentration of 200 nM each in IDT duplex buffer in a final volume of 20 μL. The gRNA complex was formed by incubation at 95°C for 5 min, followed by slowly cooling down for 20 min at room temperature, and then stored at −20°C. To form the RNP complex, 8.3 μL of HiFi Cas9 Nuclease (IDT, catalog no. 1081060), and 6 μL of the previously made gRNA was mixed in nuclease-free water to a 25-μL final volume. The mixture was incubated for 15 min at room temperature prior to use.
(ii) Confirmation of gene deletion strains.
After 5 to 7 days of incubation at 25°C, colonies grown on SMM plates supplemented with hygromycin (150 μg/mL) were subjected to a second round of selection on hygromycin. Single-spored transformants were later tested for proper homologous recombination at the ORF locus by PCR.
To confirm deletion of laeA, 25 hygromycin-resistant transformants were isolated after a rapid selection procedure on SMM supplemented with hygromycin. The correct replacement of the laeA with the hph gene was first verified by PCR analysis of genomic DNA derived from the transformant strains using primers that amplify the laeA ORF. One ORF-specific confirmation primer set (PMB_laeA_F/R) was chosen for the strain. About 40% (10/25) of the monoconidial lines generated from primary transformants of strain MB were PCR-positive for the absence of the laeA ORF (data not shown). The positive deletion strains identified by PCR were further checked for a single insertion of the deletion cassette by Southern blot analysis. Probes corresponding to the 5′ and 3′ flanks of the laeA gene in each strain were labeled using [α-32P]dCTP (Perkin-Elmer, USA) according to the manufacturer’s instructions. A single-site integration of the deletion cassette was revealed in a single transformant of the strain MB (see Fig. S1b). A HygR control strain, which has the hygromycin cassette inserted into the genome but not at the target locus, was also included in the study as a control for absence of selectable marker gene effects. To confirm deletion of psoA in Penicillium strain MB_WT using Cas9/sgRNA RNP complex, four hygromycin-resistant transformants were isolated after a rapid selection procedure on SMM supplemented with hygromycin. Two out of four of the monoconidial lines generated were PCR positive for the absence of the psoA ORF using the primer set PMB_psoA_F/R (data not shown). Later, identification of exact gene deletion locations and assessment of off-target effects of the cas9 were analyzed by whole-genome resequencing of the two PCR- positive knockout strains. DNA was extracted from each ΔpsoA strain using a Qiagen DNeasy PowerSoil extraction kit. DNA was sent to the Microbial Genome Sequencing Center for library preparation and sequencing on an Illumina NextSeq 2000. To determine the specific location of CRISPR deletion, reads were mapped to the Penicillium sp. MB_WT genome using Bowtie. To assess whether any off-target mutations were caused by the CRISPR deletion of psoA, FreeBayes was used to identify variants in the mappings of both ΔpsoA strains. Overall, only two high-confidence single nucleotide polymorphisms (SNPs) were detected in both CRISPR knockout strains that could have resulted from off-target effects: one SNP (G→A with predicted amino acid substitution of D→N) in a gene annotated as “chromatin structure-remodeling complex subunit rsc1” and another SNP (DNA G→A with predicted amino acid substitution of P→L) in a predicted protein with an unknown function (see Fig. S1g).
Construction and confirmation of complement strains.
To confirm that the phenotype exhibited by the MB ΔlaeA strain is caused by the deletion of this specific gene, the MB ΔlaeA strain was complemented with a wild-type (WT) copy of the laeA gene using phleomycin as a selectable marker. Restriction sites for NotI were introduced at the predicted native promoter and terminator of the laeA gene using primers PMB_laeAcomp_F and PMB_laeAcomp_R. The laeA gene was cloned into the pBC-phleo plasmid at the multiple cloning site located within the lacZ gene (see Fig. S1c). The ligation of the digested insert into the recipient plasmid was performed using T4 DNA ligase (New England Biolabs) following the manufacturer’s instructions. The ligation reaction was later transformed into Escherichia coli DH5α competent cells according to the manufacturer’s directions (Thermo Fisher Scientific). Five white bacterial colonies were randomly selected from the blue-white screening lysogeny broth (LB) agar plate and screened for successful ligations by conducting a diagnostic restriction digest with the NotI restriction enzyme. The E. coli strain carrying the correct plasmid (labeled PJT3) was then grown in 50 mL of LB supplemented with chloramphenicol (35 μg/mL), and the plasmid DNA was isolated using a Quantum Prep-Plasmid Midiprep kit (Bio-Rad) according to the manufacturer’s instructions. Next, 10 μg of plasmid DNA was used for the transformation of the MB_ΔlaeA strain following the same protocol described above. Prior to transformation, the plasmid was linearized using the NotI restriction enzyme.
Southern blotting was performed to confirm the single integration of laeA into the MB deletion strain using the laeA ORF sequence (amplified using the primer set laeA_ORF_F/R) for making the probe. Genomic DNAs from both laeA complement and deletion strains were digested with the same enzyme used for cloning. A positive control corresponding to the PJT3 plasmid was incorporated in the Southern blot analysis, and the MB_ΔlaeA strain was used as a negative control. 10 phleomycin-resistant transformants were isolated and subjected to Southern blot to confirm the single insertion of the laeA ORF. One strain out of 10 showed a single band of 2.8 Kb that matches the band obtained with the positive control (plasmid PJT3 generated after subcloning the laeA fragment into plasmid pBC-phleo). As expected, the MB_ΔlaeA mutant strain used as a negative control did not show any band (see Fig. S1d).
Morphophysiological analysis.
The impact of laeA deletion on the morphophysiological traits of the cheese Penicillium sp. strain MB was evaluated by monitoring the growth, sporulation, and germination of the MB_WT strain in comparison to the MB_ΔlaeA, MB_HygR, and MB_laeAc control strains. The phenotypic appearance and vegetative growth were evaluated on five different media: GMM, CYA, YES, and CCA.
Spore production and germination were assessed on GMM agar and broth, respectively. For conidial counts, fresh spores from each strain were diluted to 105 spores/mL in GMM top agar and overlaid onto agar plates of the same medium. The plates were incubated at 25°C and agar plugs removed on the second, third, and fourth day post-inoculation were homogenized in 3 mL of 0.01% Tween 80 using the VWR 200 homogenizer. Total spore counts were made using a hemocytometer. Conidial germination rates were evaluated over a 24-h growth period using a Nikon Ti inverted microscope. A spore suspension of 105 spores/mL of GMM broth was prepared for each strain, and about 1 mL was distributed into three replicate wells of a 24-sterile well plate. Five pictures per well were taken an hour apart beginning 4 h postincubation. The number of germlings were counted for each strain and the percentage of germinated spores was plotted against time to estimate the germination rates.
Community and pairwise interaction assays.
To determine how the deletion of laeA impacted microbial community assembly, we reconstructed cheese rind bacterial communities on cheese curd agar (CCA) with each of the Penicillium strains and measured total bacterial community abundance (as total CFU) and bacterial community composition (relative abundance of each community member) at 3, 10, and 21 days of community assembly. Each member of a four-member bacterial community (Staphylococcus equorum BC9, Brevibacterium auranticum JB5, Brachybacterium alimentarium JB7, and Psychrobacter sp. strain JB193) was initially inoculated at 200 CFU per species in five treatments: (i) bacteria alone, (ii) bacteria + Penicillium WT strain, (iii) bacteria + Penicillium ΔlaeA strain (MB_ΔlaeA), (iv) bacteria + Penicillium hygromycin resistance control strain (MB_HygR), and (v) bacteria + Penicillium laeA complement strain (MB_laeAc). Penicillium strains were also inoculated at 200 CFU from experimental glycerol stocks (46). For each treatment, replicate communities were inoculated on the surface of 150 μL of cheese curd agar dispensed into each well of a 96-well plate. To determine bacterial community composition, communities were harvested from individual wells with a sterile toothpick, suspended in 500 μL of phosphate-buffered saline (PBS) in a 1.5-mL microcentrifuge tube, homogenized with a sterile micropestle, and serially diluted onto plate count agar with milk and salt (PCAMS) media (46). To selectively plate bacteria, 100 mg/L of cycloheximide was added to PCAMS. To quantify Penicillium abundance, 50 mg/L of chloramphenicol was added to PCAMS. Each of the four bacteria have very distinct colony morphologies, making it easy to determine the abundance of each community member.
To determine whether the presence of another fungus could modify the inhibitory effects of Penicillium sp. strain MB, we repeated the community assays above with an isolate of the yeast Debaryomyces hansenii (see Fig. S3). This is a very widespread yeast in cheese rinds, has neutral or sometimes positive effects on the growth of cheese rind bacteria, and often co-occurs with Penicillium species (39). These experiments with the additional yeast were repeated as described above except that 200 CFU of Debaryomyces hansenii strain 135B were added to all treatments. Bacterial and fungal abundances were quantified as described above.
Pairwise interactions between each individual bacterium and the four Penicillium strains (MB_WT, MB_ΔlaeA, MB_HygR, and MB_laeAc) were assessed using the same experimental setup as the community experiments. Each bacterium was inoculated on the surface of a well of a 96-well plate with PCAMS either alone or with 200 CFU of each of the four Penicillium strains. Bacterial abundance was determined at 3, 10, and 21 days by plating harvested cocultures on PCAMS supplement with 100 mg/L of cycloheximide.
To determine the role of pseurotin in shaping cheese microbial communities, these community assays were conducted with the ΔpsoA-1 and ΔpsoA-2 strains. The experimental setup and data collection and analysis were identical to the experiments with the MB_WT, MB_ΔlaeA, MB_HygR, and MB_laeAc strains noted above.
RNA sequencing analysis and antiSMASH BGC prediction.
Transcriptome changes in Penicillium sp. strains MB_WT and MB_ΔlaeA were investigated using RNA-sequencing analysis of cultures growing on CCA medium. Inoculum of both strains were prepared from 1-week cultures on PCAMS medium. A 1-cm2 plug was taken from the leading edge of mycelium and homogenized in 500 μL of PBS. A 20 μL inoculum was spotted onto a CCA plate at three evenly spaced positions. After 48 h of growth in the dark at 24°C, the spots were about 1.5 cm in width. The MB_WT had produced blue colored spores whereas the MB_ΔlaeA spores were lighter in color. The entire fungal growth from each spot was cutoff from the CCA plates, placed in RNAlater (Qiagen), and stored at −80°C. Four biological replicates were sampled for each strain.
RNA was extracted from one of the three spots from each replicate plate using the Qiagen RNeasy Plant minikit after grinding the sample in liquid nitrogen. Approximately 100 mg of ground fungal biomass was mixed in 750 μL of Buffer RLT supplemented with 10 μL of β-mercaptoethanol. The manufacturer’s recommended protocol was followed for RNA extraction, including an on-column DNase treatment. To isolate mRNA, the NEBNext Poly(A) mRNA magnetic isolation module (New England Biolabs) was used. This mRNA was used to generate RNA-seq libraries using the NEBNext Ultra II RNA Library Prep kit for Illumina according to the manufacturer’s recommended protocol. The RNA-seq libraries were sequenced using 125-bp paired-end Illumina sequencing on a HiSeq at the Harvard Bauer Core.
Duplicate reads were removed, and the total number of reads was subsampled to 3.8 million forward reads that were used for read mapping and differential expression analysis. Reads were mapped to a draft genome of Penicillium sp. strain MB. Read mapping was performed with TopHat v2.1.0 (73). Differentially expressed genes were identified using DeSeq2 (74). Genes with a >5-fold change in expression and false discovery rate (FDR)-corrected P values of <0.05 were considered differentially expressed. To identify specific biological pathways that were enriched in the sets of downregulated or upregulated genes, we used a KOBAS 2.0 (75) to conduct a hypergeometric test on functional assignments from the Gene Ontology (GO) database (using the Aspergillus flavus genome as a reference for GO ID assignment) with Benjamini-Hochberg FDR correction. To identify putative BGCs in the Penicillium sp. strain MB genome beyond the pseurotin gene cluster, we used the fungal version of antiSMASH v 6.1.1 (76).
Metabolite profiling by UHPLC-MS analysis.
To determine the effect of laeA deletion on the biosynthetic metabolome of the Penicillium sp. strain MB, all four strains (MB_WT, MB_ΔlaeA, MB_HygR, and MB_laeAc) were cultivated by centrally inoculating 106 fresh spores on 60-mm petri dishes containing 10 mL of the agar media PDA, CYA, YES, and CCA. Three technical replicates per strain and condition were prepared. The cultures were incubated at 25°C for 2 weeks. After the incubation period, all cultures were freeze-dried (~3 g [dry weight]) and ground into 5 mL of sterile water. Soluble metabolites were later extracted by solvent extraction procedure using 5 mL of ethyl acetate. An organic metabolite fraction was generated by liquid-liquid partitioning and dried under vacuum. The crude extract was then dissolved in 400 μL acetonitrile-water (80:20 [vol/vol]) at a concentration of 100 μg/μL. Samples were later analyzed by UHPLC-MS as previously described (77). The total data set was first evaluated using the software MAVEN and the XCMS open-source package. Differential masses found via XCMS were filtered by having a maximum intensity greater than 4 × 104. Identified masses that had a maximum intensity lower than 4 × 104 were considered as background. A volcano plot was later constructed to determine statistically significant data points in crude extracts analyzed for both MB_WT and MB_ΔlaeA in negative ionization mode. For volcano plot construction, metabolites were filtered based on a P value of <0.05 and a fold change higher than 2 and lower than −2.
To confirm the lack of pseurotin production by the two ΔpsoA mutants, crude extracts from 14-day cultures on YES agar medium of both MB_ΔpsoA and MB_WT were obtained and assessed by high-resolution parallel reaction monitoring (PRM) LC-MS/MS using a Vanquish uHPLC plumbed directly to a Q-Exactive Plus mass spectrometer (Thermo Scientific) outfitted with a 75-μm ID nanospray emitter packed with 15-cm Kinetex C18 resin (1.7-μm particle size; Phenomenex). Mobile phases included solvent A (95% H2O, 5% acetonitrile, 0.1% formic acid) and solvent B (30% H2O, 70% acetonitrile, 0.1% formic acid). Nanoliter flow rates were achieved by uHPLC split-flow and measured as 300 nL/min at the nanospray emitter. First, 5 μL of each sample was autoinjected prior to the split, leading to the separation and analysis of 10 nL of extract over a 30-min chromatographic method: 0 to 100% B over 17 min; 100 to 0% B over 3 min; hold at 0% B for 10 min to re-equilibrate the column. Pseurotin A was targeted for PRM analysis in positive-ion mode with a duty cycle that included a selected ion monitoring scan (427 to 437 m/z range; resolution, 35,000; 2 microscan spectrum averaging), followed by a PRM scan targeting the pseurotin A ion (m/z of 432.1653 [M+H]+; resolution 17,500; 2.0 m/z isolation window with a 0.5 m/z offset; normalized collision energy [NCE] in the HCD cell at 35). Sample extracts were measured across three technical replicates of each strain (WT_MB, MB_ΔpsoA1, and MB_ΔpsoA2), including 10 μM pseurotin A standard (Cayman Chemical). Standard addition experiments were also performed using the above PRM method on samples extracted from YES grown MB_WT strain to assess the pseurotin A concentration. All PRM data were analyzed using Skyline (78) and FreeStyle (Thermo Scientific) software.
In vitro antimicrobial assay.
To determine whether the findings observed with community and pairwise interaction assays are due to secreted metabolite(s), the antimicrobial activities of all crude exudates collected from cultures of MB_WT, MB_ΔlaeA, MB_HygR, and MB_laeAc strains on various media were evaluated using the paper disk agar diffusion method. The antimicrobial properties were assessed against the same bacterial strains used for community and pairwise interaction assays except the B. aurantiacum strain JB5 due to the inability of growing this strain for these in vitro experiments. Bacterial strains were first cultured in 5 mL of LB broth under 280 rpm at room temperature for 24 to 48 h. The optical density of the bacterial suspension was later adjusted to 1. One milliliter of the bacterial suspension was then added to 20 mL of LB top agar, and 5 mL was gently applied on agar dishes of the same medium. In sequence, sterile disks impregnated with 10 μL of extracts (at a concentration of 100 μg/μL) dissolved in acetonitrile-water (80:20 [vol/vol]) was placed over the bacterial culture plates. One disk containing the solvent previously used for resuspension was used as negative control. For each bacterium, one disk of ampicillin at a concentration of 100 μg/mL was applied as a positive control. All dishes were incubated at room temperature, for 24 to 48 h. At the end of the incubation period, each dish was examined, and inhibition halo diameters were measured.
Comparative genomic analysis of the pseurotin gene cluster.
To contextualize the ecological importance of pseurotin in Penicillium spp. relative to other fungi, we used a data set of all annotated publicly available genomes originally downloaded from NCBI on 20 April 2020. This data set comprised 1,464 genomes representing 808 species. We performed pairwise reciprocal-best hit analysis of all proteins in the Aspergillus fumigatus genome against all 1,464 genomes using methods described previously (79). The results of this analysis were used to identify psoA, psoB, psoC, psoD, psoE, psoF, psoG, and fapR orthologs across fungal phyla.
We mapped the phylogenetic relationship of fungal genomes based on 290 benchmarking single copy orthologs (BUSCOs) as identified with BUSCO (80). We aligned sequences of each BUSCO using MAFFT and trimmed alignments using TrimAl (81) with the parameter -automated1. We constructed phylogenies for each gene using IQtree (82) after testing for the best fit model. We then created a single consensus tree using ASTRAL (83).
We selected a subset of genomes to represent taxonomic breadth based on visual inspection of phylogenetic relationships between species where seven or eight pseurotin genes were found. In addition, we added a set of genomes that were not present on NCBI but were found to contain the pseurotin gene cluster from our analysis of this cluster in the Penicillium genus (see above). Phylogenetic relationships between these genomes were determined using the same methodology as described above. Pseurotin gene clusters were determined from antiSMASH (84) by selecting cluster calls that contained the psoA ortholog (as determined above). The resulting clusters were aligned and visualized using Clinker (85). When BGC border calls made by antiSMASH extended beyond the pseurotin BGC, we trimmed these calls to facilitate visualization of this gene cluster. Validation of gene calls in genome annotations was beyond the scope of this study. To explore the evolutionary history of the pseurotin gene cluster, we aligned PsoA, PsoB, and PsoC sequences of the selected species using MAFFT (86) using the same parameters described above. The resulting alignments were trimmed with TrimAl (81) and then concatenated to form a single sequence for each species. A phylogeny was generated from this alignment using IQtree (82). Phylogenies were visualized in R (87) using ggtree (88).
Availability of biological materials.
All unique materials, including the Penicillium sp. strain MB_WT isolated from cheese, the Penicillium sp. strain MB_laeA deletion mutant, and the bacterial strains used in the interaction and antimicrobial assays, are readily available from the authors upon request.
Data availability.
Sequence data that support the findings of this study have been deposited in the NCBI SRA database with accession numbers PRJNA861320 for the whole-genome resequencing data and PRJNA861316 for the RNA-seq data. The LC-MS raw data have also been deposited to MassIVE (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp), and data are available at ftp://massive.ucsd.edu/MSV000090563/ (data set ID MSV000090563, password: cheese).
Source data used to create all figures are available in the supplemental material.
Statistical tests and exact P values for each experiment. Download Data Set S1, XLSX file, 0.6 MB (587.2KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
RNA-seq and AntiSMASH data. (a) Differentially expressed genes between MB_WT and MB_ΔlaeA strains. (b). KOBAS pathway enrichment analysis of downregulated and upregulated pathways in WT versus laeA-deleted Penicillium sp. strain MB. (c) Overview of AntiSMASH results for the genome of Penicillium sp. strain MB. Download Data Set S2, XLSX file, 0.2 MB (228.7KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Reciprocal best-hit BLAST results demonstrating the presence of the nine different genes in the pseurotin BGC across many fungal genomes. 1, gene is present. Download Data Set S3, XLSX file, 0.1 MB (119.1KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
ACKNOWLEDGMENTS
This study was supported by a grant from the National Science Foundation (CAREER 1942063) to B.E.W., a Secure Ecosystem Engineering and Design project funded by the Genomic Science Program of the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research as part of the Secure Biosystems Design Science Focus Area to P.E.A.
Ruby Ye provided feedback on the manuscript and experimental design.
Conceptualization was carried out by J.T., B.E.W., and N.P.K. Experiments were performed by J.T., C.M.C., M.T.D., and B.E.W. The article was written by J.T., C.M.C., and B.E.W. and revised with input from all authors. The figures and statistical analyses were made by J.T., C.M.C., T.A.R., and B.E.W., except for Fig. 4 (M.T.D.); see also Fig. S5 (R.J.G.), and Fig. S6 (M.T.D.). The RNA-seq experiments were conducted and analyzed by B.E.W. The LC-MS curation of data and analysis were conducted by R.J.G., P.E.A., T.A.R., and J.T. The study was supervised by N.P.K. and B.E.W.
This manuscript has been authored by UT-Battelle, LLC, under contract DE-AC05-00OR22725 with the U.S. Department of Energy (DOE).
We declare there are no competing interests.
Contributor Information
Nancy P. Keller, Email: npkeller@wisc.edu.
Benjamin E. Wolfe, Email: benjamin.wolfe@tufts.edu.
John W. Taylor, University of California—Berkeley
REFERENCES
- 1.Fleming A. 1929. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. Br J Exp Pathol 10:226. [PMC free article] [PubMed] [Google Scholar]
- 2.Peplow AW, Meek IB, Wiles MC, Phillips TD, Beremand MN. 2003. Tri16 is required for esterification of position C-8 during trichothecene mycotoxin production by Fusarium sporotrichioides. Appl Environ Microbiol 69:5935–5940. doi: 10.1128/AEM.69.10.5935-5940.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhatnagar D, Cary JW, Ehrlich K, Yu J, Cleveland TE. 2006. Understanding the genetics of regulation of aflatoxin production and Aspergillus flavus development. Mycopathologia 162:155–166. doi: 10.1007/s11046-006-0050-9. [DOI] [PubMed] [Google Scholar]
- 4.Georgianna DR, Payne GA. 2009. Genetic regulation of aflatoxin biosynthesis: from gene to genome. Fungal Genet Biol 46:113–125. doi: 10.1016/j.fgb.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 5.Gil-Serna J, Vázquez C, Patiño B. 2020. Genetic regulation of aflatoxin, ochratoxin A, trichothecene, and fumonisin biosynthesis: a review. Int Microbiol 23:89–96. doi: 10.1007/s10123-019-00084-2. [DOI] [PubMed] [Google Scholar]
- 6.Keller NP, Turner G, Bennett JW. 2005. Fungal secondary metabolism: from biochemistry to genomics. Nat Rev Microbiol 3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 7.Betina V, Mičeková D, Nemec P. 1972. Antimicrobial properties of cytochalasins and their alteration of fungal morphology. Microbiology 71:343–349. doi: 10.1099/00221287-71-2-343. [DOI] [Google Scholar]
- 8.Calvo AM, Wilson RA, Bok JW, Keller NP. 2002. Relationship between secondary metabolism and fungal development. Microbiol Mol Biol Rev 66:447–459. doi: 10.1128/MMBR.66.3.447-459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calvo AM, Cary JW. 2015. Association of fungal secondary metabolism and sclerotial biology. Front Microbiol 6:62. doi: 10.3389/fmicb.2015.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yim G, Wang HH, Davies J. 2007. Antibiotics as signaling molecules. Philos Trans R Soc Lond B Biol Sci 362:1195–1200. doi: 10.1098/rstb.2007.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maillet F, Poinsot V, André O, Puech-Pagès V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A, Martinez EA, Driguez H, Bécard G, Dénarié J. 2011. Fungal lipochito-oligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469:58–63. doi: 10.1038/nature09622. [DOI] [PubMed] [Google Scholar]
- 12.Pusztahelyi T, Holb IJ, Pócsi I. 2015. Secondary metabolites in fungus-plant interactions. Front Plant Sci 6:573. doi: 10.3389/fpls.2015.00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rush TA, Puech-Pagès V, Bascaules A, Jargeat P, Maillet F, Haouy A, Maës AQ, Carriel CC, Khokhani D, Keller-Pearson M, Tannous J, Cope KR, Garcia K, Maeda J, Johnson C, Kleven B, Choudhury QJ, Labbé J, Swift C, O’Malley MA, Bok JW, Cottaz S, Fort S, Poinsot V, Sussman MR, Lefort C, Nett J, Keller NP, Bécard G, Ané J-M. 2020. Lipo-chitooligosaccharides as regulatory signals of fungal growth and development. Nat Commun 11:3897. doi: 10.1038/s41467-020-17615-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scharf DH, Heinekamp T, Brakhage AA. 2014. Human and plant fungal pathogens: the role of secondary metabolites. PLoS Pathog 10:e1003859. doi: 10.1371/journal.ppat.1003859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Snini SP, Tannous J, Heuillard P, Bailly S, Lippi Y, Zehraoui E, Barreau C, Oswald IP, Puel O. 2016. Patulin is a cultivar-dependent aggressiveness factor favoring the colonization of apples by Penicillium expansum. Mol Plant Pathol 17:920–930. doi: 10.1111/mpp.12338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Losada L, Ajayi O, Frisvad JC, Yu J, Nierman WC. 2009. Effect of competition on the production and activity of secondary metabolites in Aspergillus species. Med Mycol 47(Suppl 1):S88–S96. doi: 10.1080/13693780802409542. [DOI] [PubMed] [Google Scholar]
- 17.Drott MT, Debenport T, Higgins SA, Buckley DH, Milgroom MG. 2019. Fitness cost of aflatoxin production in Aspergillus flavus when competing with soil microbes could maintain balancing selection. mBio 10:e02782-18. doi: 10.1128/mBio.02782-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costa JH, Wassano CI, Angolini CFF, Scherlach K, Hertweck C, Fill TP. 2019. Antifungal potential of secondary metabolites involved in the interaction between citrus pathogens. Sci Rep 9. doi: 10.1038/s41598-019-55204-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rohlfs M, Albert M, Keller NP, Kempken F. 2007. Secondary chemicals protect mould from fungivory. Biol Lett 3:523–525. doi: 10.1098/rsbl.2007.0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drott MT, Lazzaro BP, Brown DL, Carbone I, Milgroom MG. 2017. Balancing selection for aflatoxin in Aspergillus flavus is maintained through interference competition with, and fungivory by insects. Proc Biol Sci 284:20172408. doi: 10.1098/rspb.2017.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jakubczyk D, Dussart F. 2020. Selected fungal natural products with antimicrobial properties. Molecules 25:911. doi: 10.3390/molecules25040911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aly AH, Debbab A, Proksch P. 2011. Fifty years of drug discovery from fungi. Fungal Divers 50:3–19. doi: 10.1007/s13225-011-0116-y. [DOI] [Google Scholar]
- 23.Keller NP. 2019. Fungal secondary metabolism: regulation, function, and drug discovery. Nat Rev Microbiol 17:167–180. doi: 10.1038/s41579-018-0121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bayram O, Krappmann S, Ni M, Bok JW, Helmstaedt K, Valerius O, Braus-Stromeyer S, Kwon N-J, Keller NP, Yu J-H, Braus GH. 2008. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320:1504–1506. doi: 10.1126/science.1155888. [DOI] [PubMed] [Google Scholar]
- 25.Bok JW, Keller NP. 2004. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell 3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perrin RM, Fedorova ND, Bok JW, Cramer RA, Jr, Wortman JR, Kim HS, Nierman WC, Keller NP. 2007. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog 3:e50. doi: 10.1371/journal.ppat.0030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim H-K, Lee S, Jo S-M, McCormick SP, Butchko RAE, Proctor RH, Yun S-H. 2013. Functional roles of FgLaeA in controlling secondary metabolism, sexual development, and virulence in Fusarium graminearum. PLoS One 8:e68441. doi: 10.1371/journal.pone.0068441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar D, Barad S, Chen Y, Luo X, Tannous J, Dubey A, Glam Matana N, Tian S, Li B, Keller N, Prusky D. 2017. LaeA regulation of secondary metabolism modulates virulence in Penicillium expansum and is mediated by sucrose. Mol Plant Pathol 18:1150–1163. doi: 10.1111/mpp.12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lv Y, Lv A, Zhai H, Zhang S, Li L, Cai J, Hu Y. 2018. Insight into the global regulation of laeA in Aspergillus flavus based on proteomic profiling. Int J Food Microbiol 284:11–21. doi: 10.1016/j.ijfoodmicro.2018.06.024. [DOI] [PubMed] [Google Scholar]
- 30.Zhu C, Wang Y, Hu X, Lei M, Wang M, Zeng J, Li H, Liu Z, Zhou T, Yu D. 2020. Involvement of LaeA in the regulation of conidia production and stress responses in Penicillium digitatum. J Basic Microbiol 60:82–88. doi: 10.1002/jobm.201900367. [DOI] [PubMed] [Google Scholar]
- 31.Zhang M, Yang Y, Li L, Liu S, Xue X, Gao Q, Wang D, Zhang Y, Zhang J. 2022. LaeA regulates morphological development and ochratoxin A biosynthesis in Aspergillus niger. Mycotoxin Res 38:221–229. doi: 10.1007/s12550-022-00463-1. [DOI] [PubMed] [Google Scholar]
- 32.Zhao X, Spraker JE, Bok JW, Velk T, He Z-M, Keller NP. 2017. A cellular fusion cascade regulated by LaeA is required for sclerotial development in Aspergillus flavus. Front Microbiol 8:1925. doi: 10.3389/fmicb.2017.01925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knowles SL, Raja HA, Wright AJ, Lee AML, Caesar LK, Cech NB, Mead ME, Steenwyk JL, Ries LNA, Goldman GH, Rokas A, Oberlies NH. 2019. Mapping the fungal battlefield: using in situ chemistry and deletion mutants to monitor interspecific chemical interactions between fungi. Front Microbiol 10:285. doi: 10.3389/fmicb.2019.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Irlinger F, Layec S, Hélinck S, Dugat-Bony E. 2015. Cheese rind microbial communities: diversity, composition, and origin. FEMS Microbiol Lett 362:1–11. doi: 10.1093/femsle/fnu015. [DOI] [PubMed] [Google Scholar]
- 35.Ropars J, Cruaud C, Lacoste S, Dupont J. 2012. A taxonomic and ecological overview of cheese fungi. Int J Food Microbiol 155:199–210. doi: 10.1016/j.ijfoodmicro.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 36.Frisvad JC. 2014. Taxonomy, chemodiversity, and chemoconsistency of Aspergillus, Penicillium, and Talaromyces species. Front Microbiol 5:773. doi: 10.3389/fmicb.2014.00773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diblasi L, Arrighi F, Silva J, Bardón A, Cartagena E. 2015. Penicillium commune metabolic profile as a promising source of antipathogenic natural products. Nat Prod Res 29:2181–2187. doi: 10.1080/14786419.2015.1007457. [DOI] [PubMed] [Google Scholar]
- 38.Cabañes FJ, Bragulat MR, Castellá G. 2010. Ochratoxin A-producing species in the genus Penicillium. Toxins (Basel) 2:1111–1120. doi: 10.3390/toxins2051111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Casquete R, Benito MJ, Aranda E, Martín A, Hernández A, Córdoba M de G. 2021. Cyclopiazonic acid gene expression as strategy to minimizing mycotoxin contamination in cheese. Fungal Biol 125:160–165. doi: 10.1016/j.funbio.2019.06.011. [DOI] [PubMed] [Google Scholar]
- 40.Puel O, Galtier P, Oswald IP. 2010. Biosynthesis and toxicological effects of patulin. Toxins (Basel) 2:613–631. doi: 10.3390/toxins2040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martín JF, Liras P. 2017. Secondary metabolites in cheese fungi, p 293–315. In Mérillon J-M, Ramawat KG (ed), Fungal metabolites. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 42.Vallone L, Giardini A, Soncini G. 2014. Secondary metabolites from Penicillium roqueforti, a starter for the production of gorgonzola cheese. Ital J Food Saf 3:2118. doi: 10.4081/ijfs.2014.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolfe BE, Button JE, Santarelli M, Dutton RJ. 2014. Cheese rind communities provide tractable systems for in situ and in vitro studies of microbial diversity. Cell 158:422–433. doi: 10.1016/j.cell.2014.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kastman EK, Kamelamela N, Norville JW, Cosetta CM, Dutton RJ, Wolfe BE. 2016. Biotic interactions shape the ecological distributions of Staphylococcus species. mBio 7:e01157-16. doi: 10.1128/mBio.01157-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Kastman EK, Guasto JS, Wolfe BE. 2018. Fungal networks shape dynamics of bacterial dispersal and community assembly in cheese rind microbiomes. Nat Commun 9:336. doi: 10.1038/s41467-017-02522-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cosetta CM, Wolfe BE. 2020. Deconstructing and reconstructing cheese rind microbiomes for experiments in microbial ecology and evolution. Curr Protoc Microbiol 56:e95. doi: 10.1002/cpmc.95. [DOI] [PubMed] [Google Scholar]
- 47.Gillot G, Decourcelle N, Dauer G, Barbier G, Coton E, Delmail D, Mounier J. 2016. 1-Octanol, a self-inhibitor of spore germination in Penicillium camemberti. Food Microbiol 57:1–7. doi: 10.1016/j.fm.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 48.Osbourn A. 2010. Secondary metabolic gene clusters: evolutionary toolkits for chemical innovation. Trends Genet 26:449–457. doi: 10.1016/j.tig.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 49.Wiemann P, Guo C-J, Palmer JM, Sekonyela R, Wang CCC, Keller NP. 2013. Prototype of an intertwined secondary-metabolite supercluster. Proc Natl Acad Sci USA 110:17065–17070. doi: 10.1073/pnas.1313258110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pinheiro EAA, Carvalho JM, dos Santos DCP, de Oliveria Feitosa A, Marinho PSB, Guilhon GMSP, de Souza ADL, da Silva FMA, do R Marinho AM. 2013. Antibacterial activity of alkaloids produced by endophytic fungus Aspergillus sp. EJC08 isolated from medical plant Bauhinia guianensis. Nat Prod Res 27:1633–1638. doi: 10.1080/14786419.2012.750316. [DOI] [PubMed] [Google Scholar]
- 51.Tsunematsu Y, Fukutomi M, Saruwatari T, Noguchi H, Hotta K, Tang Y, Watanabe K. 2014. Elucidation of pseurotin biosynthetic pathway points to trans-acting C-methyltransferase: generation of chemical diversity. Angew Chem 126:8615–8619. doi: 10.1002/ange.201404804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Samanidou V, Michaelidou K, Kabir A, Furton KG. 2017. Fabric phase sorptive extraction of selected penicillin antibiotic residues from intact milk followed by high performance liquid chromatography with diode array detection. Food Chem 224:131–138. doi: 10.1016/j.foodchem.2016.12.024. [DOI] [PubMed] [Google Scholar]
- 53.Fisch KM. 2013. Biosynthesis of natural products by microbial iterative hybrid PKS–NRPS. RSC Adv 3:18228–18247. doi: 10.1039/c3ra42661k. [DOI] [Google Scholar]
- 54.Breitenstein W, Chexal KK, Mohr P, Tamm C. 1981. Pseurotin B, C, D, and E: further new metabolites of Pseudeurotium ovalis STOLK. Helv Chim Acta 64:379–388. doi: 10.1002/hlca.19810640203. [DOI] [Google Scholar]
- 55.Martín JF. 2017. Key role of LaeA and velvet complex proteins on expression of β-lactam and PR-toxin genes in Penicillium chrysogenum: cross-talk regulation of secondary metabolite pathways. J Ind Microbiol Biotechnol 44:525–535. doi: 10.1007/s10295-016-1830-y. [DOI] [PubMed] [Google Scholar]
- 56.Wang G, Zhang H, Wang Y, Liu F, Li E, Ma J, Yang B, Zhang C, Li L, Liu Y. 2019. Requirement of LaeA, VeA, and VelB on asexual development, ochratoxin A biosynthesis, and fungal virulence in Aspergillus ochraceus. Front Microbiol 10:2759. doi: 10.3389/fmicb.2019.02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perlatti B, Lan N, Jiang Y, An Z, Bills G. 2020. Identification of secondary metabolites from Aspergillus pachycristatus by untargeted UPLC-ESI-HRMS/MS and genome mining. Molecules 25:913. doi: 10.3390/molecules25040913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ishikawa M, Ninomiya T, Akabane H, Kushida N, Tsujiuchi G, Ohyama M, Gomi S, Shito K, Murata T. 2009. Pseurotin A and its analogues as inhibitors of immunoglobuline E production. Bioorg Med Chem Lett 19:1457–1460. doi: 10.1016/j.bmcl.2009.01.029. [DOI] [PubMed] [Google Scholar]
- 59.Mehedi M, Molla AH, Khondkar P, Sultana S. 2010. Pseurotin A: an antibacterial secondary metabolite from Aspergillus fumigatus. Asian J Chem 22:2611–2614. [Google Scholar]
- 60.Hayashi A, Fujioka S, Nukina M, Kawano T, Shimada A, Kimura Y. 2007. Fumiquinones A and B, nematicidal quinones produced by Aspergillus fumigatus. Biosci Biotechnol Biochem 71:1697–1702. doi: 10.1271/bbb.70110. [DOI] [PubMed] [Google Scholar]
- 61.Sbaraini N, Phan C-S, Silva e Souza E, Perin APA, Rezaee H, Geremia F, da Silva Camargo M, Barbosa EG, Schrank A, Chooi Y-H, Staats CC. 2022. Intra-hemocoel injection of pseurotin A from Metarhizium anisopliae, induces dose-dependent reversible paralysis in the Greater Wax Moth (Galleria mellonella). Fungal Genet Biol 159:103675. doi: 10.1016/j.fgb.2022.103675. [DOI] [PubMed] [Google Scholar]
- 62.Martínez-Luis S, Cherigo L, Arnold E, Spadafora C, Gerwick WH, Cubilla-Rios L. 2012. Antiparasitic and anticancer constituents of the endophytic fungus Aspergillus sp. strain F1544. Nat Prod Commun 7:165–168. [PubMed] [Google Scholar]
- 63.Maiya S, Grundmann A, Li X, Li S-M, Turner G. 2007. Identification of a hybrid PKS/NRPS required for pseurotin A biosynthesis in the human pathogen Aspergillus fumigatus. Chembiochem 8:1736–1743. doi: 10.1002/cbic.200700202. [DOI] [PubMed] [Google Scholar]
- 64.Bertinetti BV, Peña NI, Cabrera GM. 2009. An antifungal tetrapeptide from the culture of Penicillium canescens. Chem Biodivers 6:1178–1184. doi: 10.1002/cbdv.200800336. [DOI] [PubMed] [Google Scholar]
- 65.Martín JF, Coton M. 2017. Blue cheese: microbiota and fungal metabolites, p 275–303. In Frias J, Martinez-Villaluenga C, Peñas E (ed), Fermented foods in health and disease prevention. Academic Press, Boston, MA. [Google Scholar]
- 66.Kure CF, Skaar I. 2019. The fungal problem in cheese industry. Curr Opin Food Sci 29:14–19. doi: 10.1016/j.cofs.2019.07.003. [DOI] [Google Scholar]
- 67.Cleary JL, Kolachina S, Wolfe BE, Sanchez LM. 2018. Coproporphyrin III produced by the bacterium Glutamicibacter arilaitensis binds zinc and is upregulated by fungi in cheese rinds. mSystems 3:e00036-18. doi: 10.1128/mSystems.00036-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pierce EC, Morin M, Little JC, Liu RB, Tannous J, Keller NP, Pogliano K, Wolfe BE, Sanchez LM, Dutton RJ. 2020. Bacterial-fungal interactions revealed by genome-wide analysis of bacterial mutant fitness. Nat Microbiol 6:87–102. doi: 10.1038/s41564-020-00800-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bodinaku I, Shaffer J, Connors AB, Steenwyk JL, Biango-Daniels MN, Kastman EK, Rokas A, Robbat A, Wolfe BE. 2019. Rapid phenotypic and metabolomic domestication of wild penicillium molds on cheese. mBio 10:e02445-19. doi: 10.1128/mBio.02445-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bok JW, Keller NP. 2012. Fast and easy method for construction of plasmid vectors using modified quick-change mutagenesis. Methods Mol Biol 944:163–174. doi: 10.1007/978-1-62703-122-6_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Niccum BA, Kastman EK, Kfoury N, Robbat A, Jr, Wolfe BE. 2020. Strain-level diversity impacts cheese rind microbiome assembly and function. mSystems 5:e00149-20. doi: 10.1128/mSystems.00149-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hassan MM, Chowdhury AK, Islam T. 2021. In silico analysis of gRNA secondary structure to predict its efficacy for plant genome editing, p 15–22. In Islam MT, Molla KA (ed), CRISPR-Cas methods, vol 2. Springer US, New York, NY. [Google Scholar]
- 73.Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li C-Y, Wei L. 2011. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blin K, Shaw S, Kloosterman AM, Charlop-Powers Z, van Wezel GP, Medema MH, Weber T. 2021. antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res 49:W29–W35. doi: 10.1093/nar/gkab335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tannous J, Kumar D, Sela N, Sionov E, Prusky D, Keller NP. 2018. Fungal attack and host defence pathways unveiled in near-avirulent interactions of Penicillium expansum creA mutants on apples. Mol Plant Pathol 19:2635–2650. doi: 10.1111/mpp.12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. 2010. Skyline: an open-source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Drott MT, Bastos RW, Rokas A, Ries LNA, Gabaldón T, Goldman GH, Keller NP, Greco C. 2020. Diversity of secondary metabolism in Aspergillus nidulans clinical isolates. mSphere 5:e00156-20. doi: 10.1128/mSphere.00156-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 81.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mirarab S, Warnow T. 2015. ASTRAL-II: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics 31:i44–52. doi: 10.1093/bioinformatics/btv234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T. 2019. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gilchrist CLM, Chooi Y-H. 2021. Clinker and clustermap.js: automatic generation of gene cluster comparison figures. Bioinformatics 37:2473–2475. doi: 10.1093/bioinformatics/btab007. [DOI] [PubMed] [Google Scholar]
- 86.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bunn A, Korpela M. 2014. Crossdating in dplR. https://cran.microsoft.com/snapshot/2014-09-08/web/packages/dplR/vignettes/xdate-dplR.pdf. Accessed 4 October 2022.
- 88.Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y. 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genetic modifications conducted in Penicillium sp. strain MB. (a) Schematic representation of the genetic construct for laeA deletion in strain MB. The construct is constituted of a gene conferring resistance to hygromycin under TrpC promoter and terminator. (b) Southern blot analyzes of genomic DNA from the WT, the ΔlaeA, and HygR strains. The positions of the restriction enzyme cutting sites used for the Southern blot are shown on the construct schematic. Portions (10 μg) of total DNA from each strain were digested with the appropriate enzymes and subjected to Southern blot analysis using, respectively, the 5′ flank fragment (blue) and the 3′ fragment (grey) as probes. The 1-kb DNA ladder from New England Biolabs was used to determine the size of the expected bands. (c) Restriction map of plasmid pBC-phleo containing the ble gene for phleomycin resistance controlled by the Aspergillus nidulans gpdA promoter and the Saccharomyces cerevisiae CYC1 terminator. Restriction site used for cloning of the laeA gene is shown in red. (d) Southern confirmation of the single insertion of laeA using NotI as a restriction enzyme. The positive control corresponds to the digested PJT3 plasmid. (e) Map of plasmid pCS01 from which the hygromycin deletion cassette was amplified and used to knock-out the psoA gene in Penicillium sp. strain MB. (f) Scheme of the experimental strategy adopted to knockout the psoA gene in the Penicillium sp. strain MB using CRISPR-Cas9 system. The CRISPR components were delivered to the strain MB_WT protoplasts as a ribonucleoprotein (RNP) complex consisting of the Cas9 nuclease and single-guide RNA (sgRNA) targeting the PKS/NRPS conserved domain in the psoA gene sequence. The RNP complex will bind to the target sequence at the specific site and cut DNA double strands. The DNA double-strand breaks will be repaired by homology directed repair using the donor DNA template containing 1 kb of homology arms flanking the hph gene that confers resistance to hygromycin. (g) Resequencing the genomes of the ΔpsoA strains revealed the deleted regions by the CRISPR-Cas9-mediated knockout. Read mappings for the two different mutants are shown, and the deleted regions are represented by the large gap in both mappings. DNA resequencing reads are indicated by black lines. Download FIG S1, TIF file, 4.8 MB (4.8MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Growth rates of Penicillium spp. on YES, PDA, CYA, and CCA. (a to c) Growth on YES medium (a), growth on PDA medium (b), and growth on CYA medium (c) are represented. In the box plots, the bar represents the standard errors of the means, and each dot represents a biological replication (n = 3). One-way analysis of variance (ANOVA) was performed for each bacterial community. Dunnett’s multiple-comparison test was used, and the results were compared to the WT strain. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; no asterisk indicates not significant (ns). For the exact P values for each treatment, see Data Set 1. (d) Growth on CCA, where points and connecting lines with standard errors of the means bars were used from two independent experiments with five biological replications each. In experiment 2, replication 5 was removed due to no growth of the bacteria in the control. Two-way ANOVA was performed for each fungal strain. Dunnett’s multiple-comparison test was used and compared to the WT strain. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; no asterisk indicates not significant (ns). For the exact P values for each treatment, see Data Set 1. Download FIG S2, TIF file, 0.9 MB (886.2KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Community analysis in the presence of another common cheese rind fungal community member, the yeast Debaryomyces hansenii. Histograms show the shifts in a typical rind community (Staphylococcus, Brevivacterium, Brachybacterium, and Psychrobacter) in the presence Penicillium sp. strain MB_WT and mutants (ΔlaeA, HygR, and laeAc) and D. hansenii after 3 (a), 10 (c), and 21 (e) days of incubation. The data are mean relative abundances from two independent experiments. Histograms show the inhibition of community members grown in the presence Penicillium sp. strain MB_WT, mutant strains, and D. hansenii after 3 (b), 10 (d), and 21 (f) days of incubation. The data are from two independent experiments, with five biological replications each. In the box plots, the bar represents the standard errors of the means, and each dot represents a biological replication. One-way ANOVA was performed for each bacterial community. Dunnett’s multiple-comparison test was used and compared to the WT. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; no asterisk indicates not significant (ns). For the exact P values for each treatment, see Data Set 1. Download FIG S3, TIF file, 1.8 MB (1.8MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Antibacterial activities of filtered crude extracts from cultures on different media. Crude extracts were collected from cultures of the different Penicillium strains on YES (a), PDA (b), CYA (c), and CCA (d) media. The diameter of the zone of inhibition was measured and analyzed. In the box plots, the bar represents the standard errors of the means, and each dot represents a biological replication (n = 3). One-way ANOVA was performed for each bacterial community. Dunnett’s multiple-comparison test was used and compared to the WT strain. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; no asterisk indicates not significant (ns). For the exact P values for each treatment, see Data Set 1. Download FIG S4, TIF file, 0.9 MB (916.3KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
LC-MS data showing the absence of pseurotin production in the crude extracts of both MB_ΔpsoA strains compared to the MB_WT strain. (a) Chromatograms showing the absence of pseurotin A and putative pseurotin D peaks in the ΔpsoA mutants. (b) Fragmentation mass spectrum in positive ionization mode of pseurotin A in MB_WT strain compared to pseurotin A standard. (c) Fragmentation mass spectrum in positive ionization mode putative pseurotin D in MB_WT strain. The MS/MS spectrum shown for the putative pseurotin D is for reference purposes only and represents the differentially eluting isomer that was also affected by the genetic knockouts performed here. Download FIG S5, TIF file, 0.8 MB (859.3KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Frequency of genes associated with the pseurotin gene cluster (left) across fungal phyla (right). A total of 1.463 fungal genomes representing 808 species were obtained from the NCBI. Phylogenetic relationships between genomes were determined from the consensus of 290 maximum-likelihood trees constructed for single-copy orthologs (BUSCOs). Terminal branch lengths were not calculated. The number of pseurotin genes present in each genome was determined using reciprocal best-hit BLAST analysis using the Aspergillus fumigatus pseurotin genes psoA, psoB, psoC, psoD, psoE, psoF, psoG, and fapR. The orange arrow indicates a monophyletic clade within Ascomycota that was visually identified as having a higher prevalence of pseurotin genes. Download FIG S6, TIF file, 0.9 MB (873.6KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Primers used in this study for the construction of deletion cassettes and molecular cloning. Download Table S1, DOCX file, 0.02 MB (16.1KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Statistical tests and exact P values for each experiment. Download Data Set S1, XLSX file, 0.6 MB (587.2KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
RNA-seq and AntiSMASH data. (a) Differentially expressed genes between MB_WT and MB_ΔlaeA strains. (b). KOBAS pathway enrichment analysis of downregulated and upregulated pathways in WT versus laeA-deleted Penicillium sp. strain MB. (c) Overview of AntiSMASH results for the genome of Penicillium sp. strain MB. Download Data Set S2, XLSX file, 0.2 MB (228.7KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Reciprocal best-hit BLAST results demonstrating the presence of the nine different genes in the pseurotin BGC across many fungal genomes. 1, gene is present. Download Data Set S3, XLSX file, 0.1 MB (119.1KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
Sequence data that support the findings of this study have been deposited in the NCBI SRA database with accession numbers PRJNA861320 for the whole-genome resequencing data and PRJNA861316 for the RNA-seq data. The LC-MS raw data have also been deposited to MassIVE (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp), and data are available at ftp://massive.ucsd.edu/MSV000090563/ (data set ID MSV000090563, password: cheese).
Source data used to create all figures are available in the supplemental material.
Statistical tests and exact P values for each experiment. Download Data Set S1, XLSX file, 0.6 MB (587.2KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
RNA-seq and AntiSMASH data. (a) Differentially expressed genes between MB_WT and MB_ΔlaeA strains. (b). KOBAS pathway enrichment analysis of downregulated and upregulated pathways in WT versus laeA-deleted Penicillium sp. strain MB. (c) Overview of AntiSMASH results for the genome of Penicillium sp. strain MB. Download Data Set S2, XLSX file, 0.2 MB (228.7KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Reciprocal best-hit BLAST results demonstrating the presence of the nine different genes in the pseurotin BGC across many fungal genomes. 1, gene is present. Download Data Set S3, XLSX file, 0.1 MB (119.1KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.