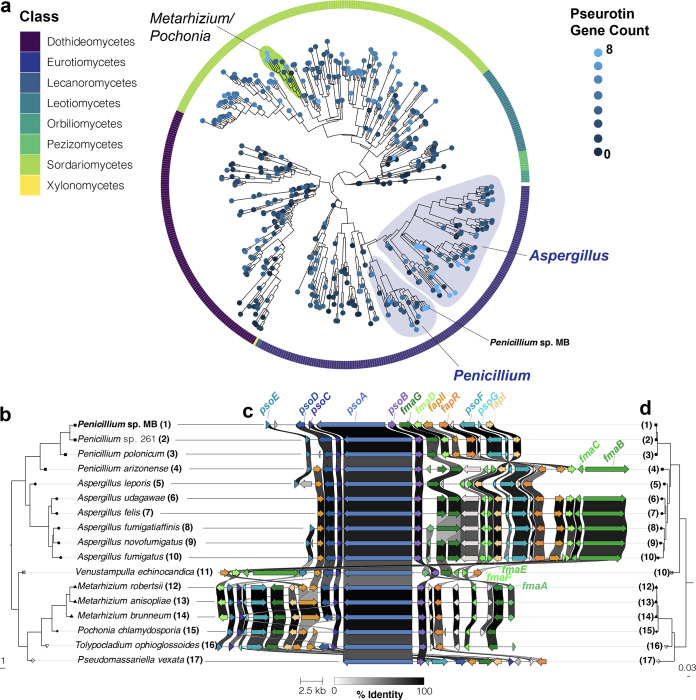
FIG 5.
Components of the pseurotin biosynthetic gene cluster are widespread across Ascomycota, but the full cluster is mainly found in Aspergillus and Penicillium species. (a) The subclade of Ascomycota identified as having higher prevalence of pseurotin cluster genes (Fig. S6) presented with a single leaf per species. Phylogenetic relationships were determined from the consensus of 290 maximum-likelihood trees constructed for benchmarking single copy orthologs (BUSCOs). The outer ring indicates the taxonomic order that species pertain to. Terminal branch lengths are not calculated. The number of pseurotin genes was determined using reciprocal best-hit BLAST analysis using the Aspergillus fumigatus pseurotin genes psoA, psoB, psoC, psoD, psoE, psoF, psoG, and fapR is indicated as the intensity of the tip color. (b) A phylogeny constructed in the same way as (a) representing a subset of species identified as having seven or eight pseurotin genes that were manually selected to represent taxonomic breadth. (c) An alignment of pseurotin gene clusters. Genes in the pseurotin cluster are labeled in blue while constituents of the intertwined fumagillin cluster are indicated in green. Regulatory genes of these clusters are labeled in orange. Coloration of genes is based on homology searches and visualizations performed by clinker. The weight of lines between homologous genes indicates the percent identity; genes sharing identity below 30% are not indicated. (d) A maximum likelihood phylogeny constructed from the concatenation of PsoA, PsoB, and PsoC sequences. Note that phylogenetic relationships between (d) and (b) are compatible, suggesting vertical transmission of the pseurotin gene cluster. The strain information of the species used in this analysis are provided in Data Set S3.