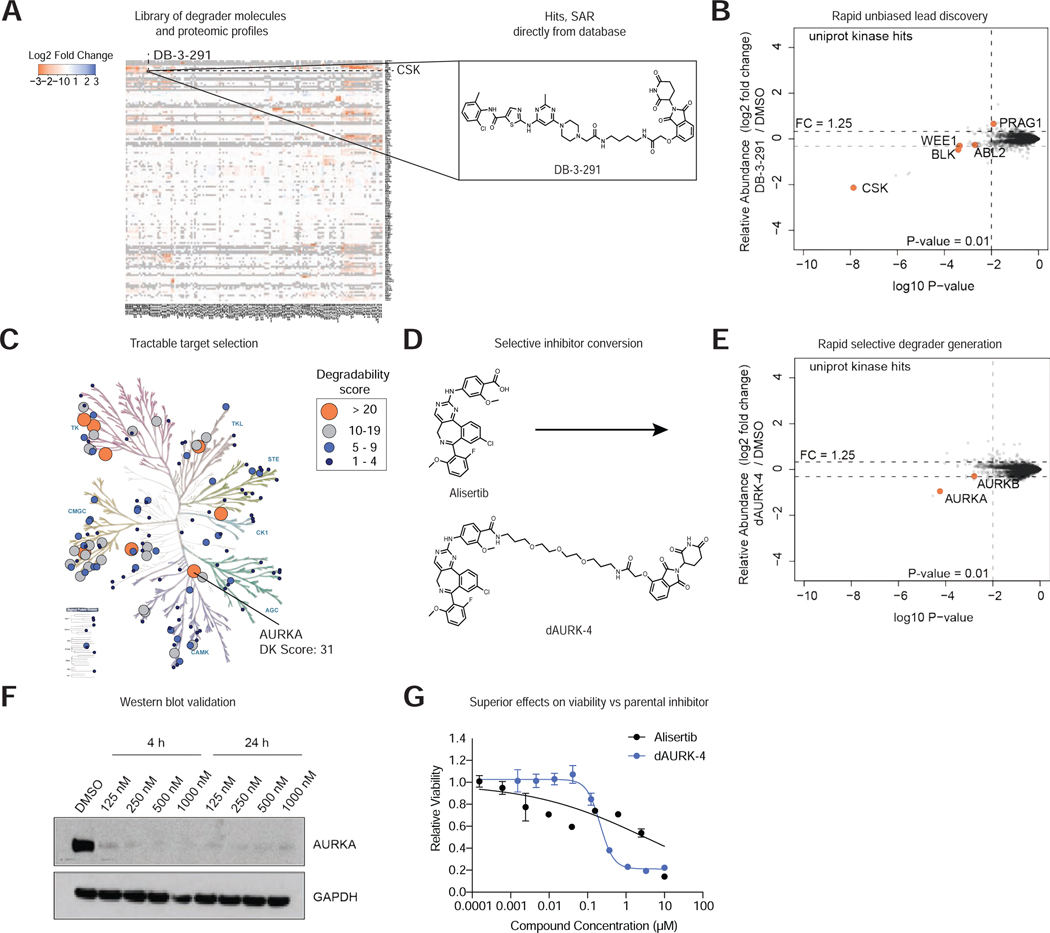
Figure 2 |. Degradable Kinome Dataset Accelerates Lead Discovery.
(A) Heatmap comparing relative fold change in protein abundance in response to treatment with indicated degrader (see Table S1 for treatment details and Table S3 for data). Inset, chemical structure of degrader DB-3-291. (B) Scatterplot displaying relative fold change in protein abundance following treatment of MOLT-4 cells with 1 μM DB-3-291 for 5 h. (C) Kinome tree representing the kinase degradability (DK) score calculated for each of the protein kinases degraded in this study. Data reported in Table S8. Image created using KinMap, illustration reproduced courtesy of Cell Signaling Technology, Inc. (www.cellsignal.com). (D) Strategy for conversion of Alisertib into selective AURKA degrader dAURK-4. (E) Scatterplot depicting relative fold change in protein abundance following treatment of MOLT-4 cells with 1 μM dAURK-4 for 5 h. Data in B, E are from n = 1 biologically independent treatment samples. Associated dataset is provided in Table S3. (F) Immunoblot analysis of MM.1S cells treated with the indicated concentration of dAURK-4 for 4 or 24 h. Data in F are representative of n = 2 independent experiments. (G) DMSO-normalized antiproliferation of MM.1S cells treated with Alisertib or dAURK-4. Data are presented as mean ± s.d. of n = 3 biologically independent samples and are representative of n = 2 independent experiments.