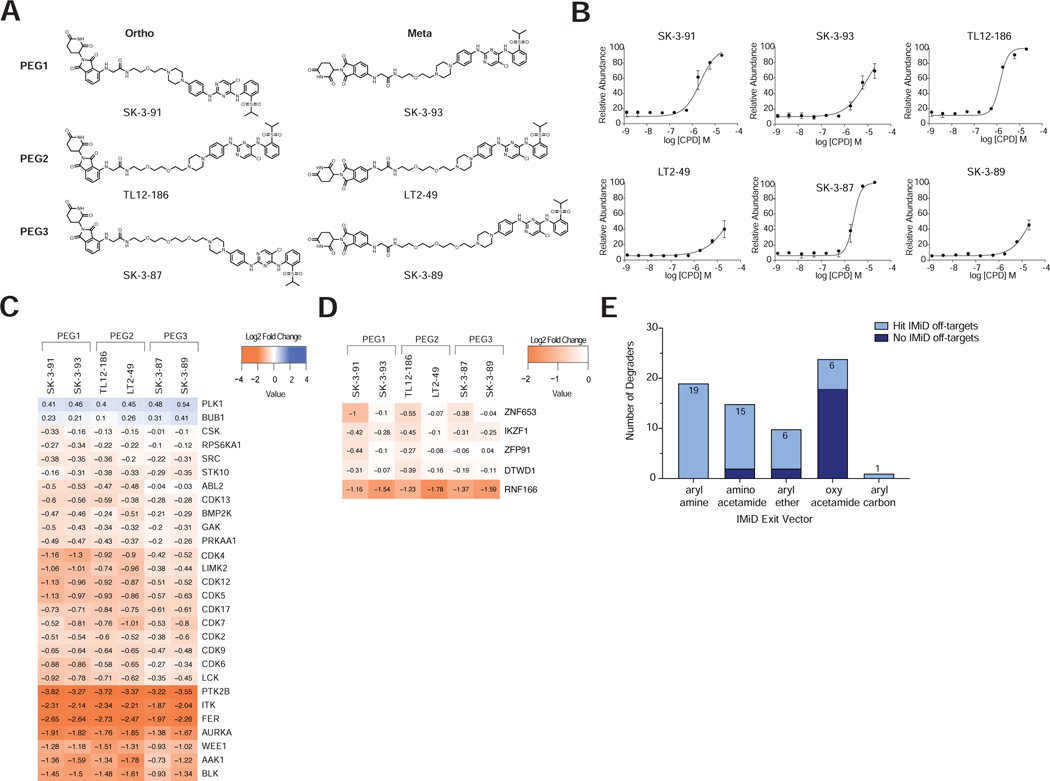
Figure 6 |. Protein kinases and IMiD off-target proteins have varied tolerance for subtle changes in linker design.
(A) Chemical structures of the compounds evaluated. (B) Intracellular ligase engagement assay. Data are represented as means ± s.d of n = 3 biologically independent replicates. (C, D) Heatmap showing log2 FC of (C) kinases, (D) known IMiD targets determined to be hits (FC >1.25 and P-value <0.01) following a 5 h treatment of MOLT-4 cells with 0.1 μM of the indicated compounds. (E) Split bar plot showing the number of CRBN-recruiting degraders found to hit at least one known IMiD off-target compared to the number that do not hit IMiD off-targets. CRBN-recruiting degraders are categorized according their linker attachment chemistry. Associated dataset is provided in Table S10.