FIG 1.
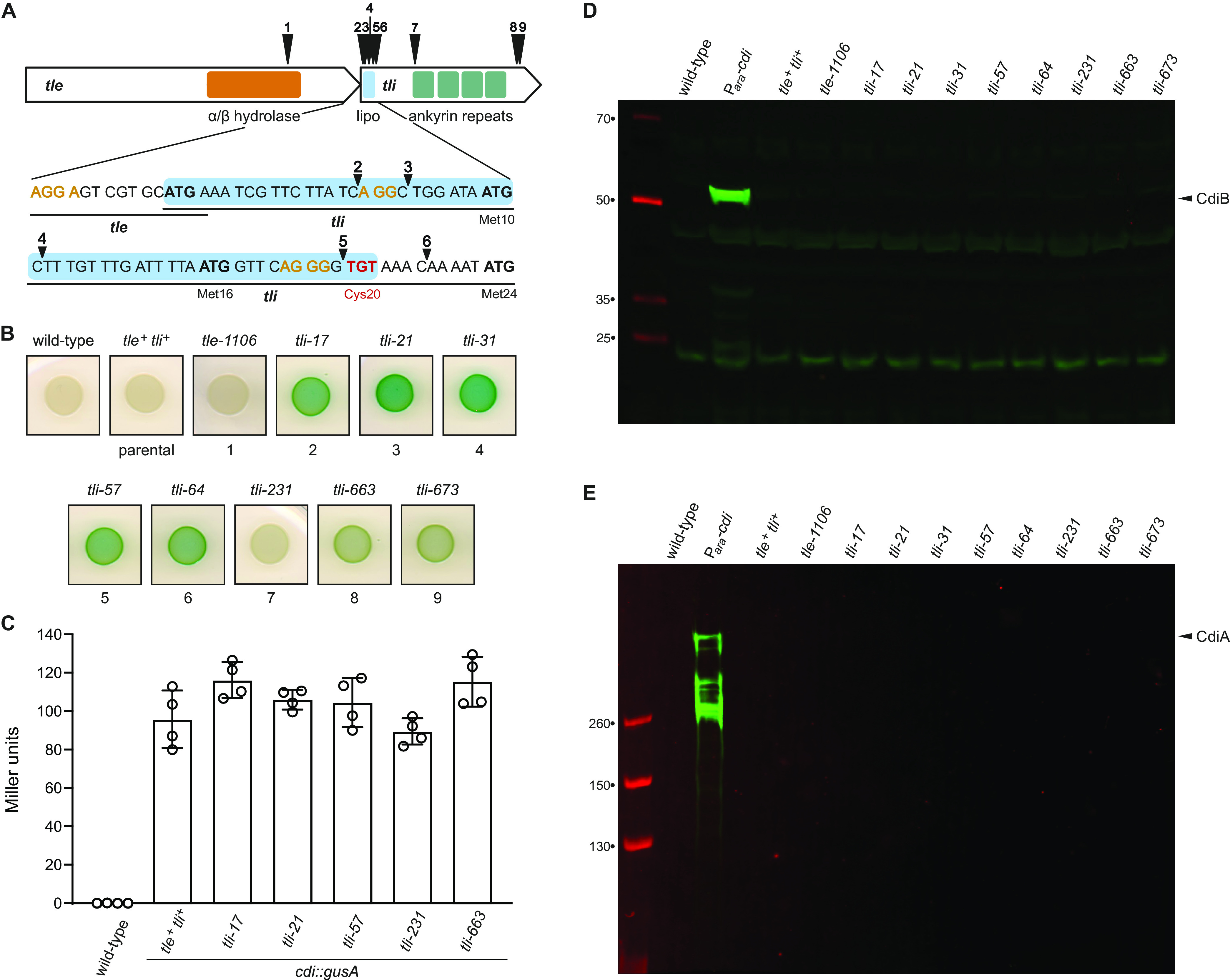
Identification of Tn5 insertions in the tli immunity gene. (A) Schematic of Tn5 insertion sites in tle and tli. Regions encoding the predicted α/β-hydrolase domain of Tle and the Tli lipoprotein signal peptide (lipo) and ankyrin repeats are depicted. Alternative translation initiation codons (Met10, Met16, and Met24) are indicated, and possible ribosome-binding sites are shown in orange font. The codon for the lipidated Cys20 residue is shown in red font. (B) Wild-type ECL, the parental cdi-gusA reporter strain (tle+ tli+), and tli mutants were grown on LB agar supplemented with X-Gluc. (C) Quantification of β-glucuronidase activity in selected tli mutants. Activities were quantified as Miller units as described in Materials and Methods. Data for technical replicates from two independent experiments are shown with the average ± standard deviation. (D and E) Tn5 insertions do not upregulate CdiB or CdiA production. Protein samples from wild-type ECL, the parental tle+ tli+ reporter strain, and tli mutants were subjected to immunoblot analysis using polyclonal antisera to CdiB (D) and CdiA (E). The Para-cdi sample is from an ECL strain that contains an arabinose-inducible promoter inserted upstream of the native cdiBAI gene cluster.
