ABSTRACT
SigS is the sole extracytoplasmic function sigma factor in Staphylococcus aureus and is necessary for virulence, immune evasion, and adaptation to toxic chemicals and environmental stressors. Despite the contribution of SigS to a myriad of critical phenotypes, the downstream effectors of SigS-dependent pathogenesis, immune evasion, and stress adaptation remain elusive. To address this knowledge gap, we analyzed the S. aureus transcriptome following transient overexpression of SigS. We identified a bicistronic transcript, upregulated 1,000-fold, containing two midsized genes, each containing single domains of unknown function (DUFs). We renamed these genes SigS-regulated orfA (sroA) and SigS-regulated orfB (sroB). We demonstrated that SigS regulation of the sroAB operon is direct by using in vitro transcription analysis. Using Northern blot analysis, we also demonstrated that SroA and SroB have opposing autoregulatory functions on the transcriptional architecture of the sigS locus, with SroA stimulating SigS mRNA levels and SroB stimulating s750 (SigS antisense) levels. We hypothesized that these opposing regulatory effects were due to a direct interaction. We subsequently demonstrated a direct interaction between SroA and SroB using an in vivo surrogate genetics approach via bacterial adenylate cyclase-based two-hybrid (BACTH) analysis. We demonstrated that the SroA effect on SigS is at the posttranscriptional level of mRNA stability, highlighting a mechanism likely used by S. aureus to tightly control SigS levels. Finally, we demonstrate that the sroAB locus promotes virulence in a murine pneumonia model of infection.
IMPORTANCE SigS is necessary for S. aureus virulence, immune evasion, and adaptation to chemical and environmental stressors. These processes are critically important for the ability of S. aureus to cause disease. However, the SigS-dependent transcriptome has not been identified, hindering our ability to identify downstream effectors of SigS that contribute to these pathogenic and adaptive phenotypes. Here, we identify a regulatory protein pair that is a major direct target of SigS, known as SroA and SroB. SroA also acts to stimulate SigS expression at the posttranscriptional level of RNA turnover, providing insight into intrinsically low levels of SigS. The discovery of SroA and SroB increases our understanding of SigS and the S. aureus pathogenesis process.
KEYWORDS: Staphylococcus aureus, sigma factors, extracytoplasmic function sigma factor, ECF, domain of unknown function, mRNA, SigS, mRNA stability
INTRODUCTION
Staphylococcus aureus has a large repertoire of virulence factors that facilitate the pathogenic process (1). Regulating the expression of virulence genes is critical for S. aureus stress adaptation and evasion of the host response, both of which are necessary for the ability of S. aureus to establish a productive infection (1). S. aureus virulence gene regulators include staphylococcal accessory regulator (Sar) proteins, RNAIII, and a host of two-component signal transduction systems (1). In addition, the activity of alternative sigma factors in S. aureus contributes significantly to virulence gene regulation by rapidly rewiring transcription in response to stressors or environmental signals.
S. aureus has three alternative sigma factors, SigB, SigH, and SigS (2–4). SigB is the general stress/stationary-phase sigma factor (5–7) that is tightly regulated through an interaction with its anti-sigma factor, RsbU (3, 8). The SigB regulon is relatively large and includes a host of critical virulence factor proteins and small RNAs (sRNAs) (SbrA, SbrB, SbrC, and Teg49) (9–15). SigH is the homologue for the competence-mediating sigma factor in Bacillus subtilis (16). The SigH regulon includes structural proteins necessary for DNA binding and uptake (2) and has been reported to induce a transient competence state and prophage excision (17–19). In this work, we focused on further defining the role of SigS.
SigS is the sole S. aureus extracytoplasmic function (ECF) sigma factor and is necessary for stress adaptation and immune evasion (4, 20). In the absence of sigS, S. aureus is attenuated for virulence and defective in producing joint inflammation in a mouse model of septic arthritis (4). S. aureus long-term viability in vitro and survival during extreme heat shock (>55°C) both require SigS (4). Finally, S. aureus sigS mutants have increased sensitivity to cell wall-targeting antibiotics, detergents, DNA-damaging agents, and innate immune system components (4, 21). Yet the mechanism whereby SigS senses these stressors and promotes adaptive phenotypes is not clear. There appears to be some continuity between these adaptive phenotypes and the induction of sigS expression, as exposure to the DNA-damaging agent methyl methanesulfonate (MMS), hydrogen peroxide (H2O2), sodium hydroxide (NaOH), immune cell components, and macrophage phagocytosis all stimulate expression of SigS (21). However, the regulation of SigS is complex, as basal expression is low in the absence of inducing signals, and it has 4 promoters controlling its transcriptional initiation (4, 21, 22). Genetic and biochemical screens identified CymR and KdpE as direct regulators, and ArlR and LacR as indirect regulators, of sigS transcription (23). Prior to this work, little was known about the posttranscriptional regulation of sigS expression or the direct targets of SigS that could promote virulence and stress adaptation.
ECF sigma factors rapidly rewire the bacterial transcriptome in response to extracytoplasmic stress (24–28). In Escherichia coli and other enteric bacteria, the ECF sigma factor, σE, has a relatively large regulon composed of periplasmic proteases and small RNAs that restore envelope homeostasis through fine-tuning of outer membrane protein levels (24–28). In B. subtilis, Clostridioides difficile, and other Firmicutes, ECF sigma factors also have relatively large regulons that ensure envelope homeostasis in the presence of cell wall stressors (29–32). Based on the function of ECF sigma factors in phylogenetically distant and related bacterial species, we hypothesized that the SigS would mediate stress adaptation, virulence, and immune evasion through the action of downstream effector proteins or sRNAs that comprise its yet-undefined direct regulon.
In this work, we analyzed the S. aureus transcriptome following transient sigS overexpression. Consequently, we identified a major SigS effector locus with a previously uncharacterized regulatory protein pair tandemly encoded on a bicistronic transcript (encoding SACOL0677 and SACOL0676). Each of these open reading frames (ORFs) is comprised of a single domain of unknown function (DUF), DUF1659 and DUF2922. We renamed these genes (SigS-regulated orfA) and orfB (sroAB), respectively. Further, we show that SroA and SroB have opposing regulatory effects on the sigS locus. We also uncovered positive feedback regulation, as SroA acts to stabilize the sigS transcript, thereby promoting SigS accumulation in the absence of SroB. Finally, we demonstrate that the sroAB locus is necessary for full virulence in a murine model of pneumonia. This work provides insight into a network that tightly controls levels of the cryptic SigS sigma factor in S. aureus.
RESULTS
S. aureus transcriptome architecture following SigS overexpression.
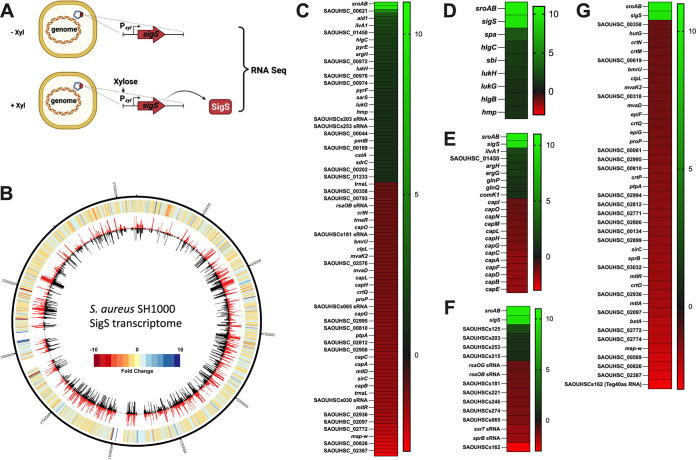
To identify downstream effectors of SigS that may promote virulence, immune evasion, or antibiotic resistance, we analyzed the S. aureus transcriptome following SigS overexpression (Fig. 1A). Briefly, exponentially growing S. aureus cultures were induced for 30 min before RNA isolation and RNA sequencing (Fig. 1A). Approximately 50 genes were upregulated and 75 genes were downregulated in S. aureus SH1000 at least 2-fold following transient overexpression of SigS (Fig. 1B and C and see Table S2 in the supplemental material). Interestingly, seven of the SigS upregulated transcripts are involved in immune evasion and coregulated by SaeR or RNAIII, including staphylococcal protein A (spa), leukocidin S subunit (hlgC), leukocidin F subunit (hlgB), IgG binding protein (sbi), gamma-hemolysin subunit (lukA), gamma-hemolysin subunit B (lukB), and nitric oxide dioxygenase (hmp) (Fig. 1D and Table S3). Seven of the SigS stimulatory targets play a role in amino acid transport or metabolism, including alanine dehydrogenase (ald1), threonine dehydratase (ilvA1), argininosuccinate lyase (argH), argininosuccinate synthase (argG), glutamate permease (glnP), glutamate ABC transporter ATP binding protein (glnQ), and a hypothetical protein, SAOUHSC_1450 (Fig. 1E and Table S4). These seven stimulatory targets of SigS are also coregulated by either GTP/branched-chained amino acid sensing global regulator (CodY), arginine repressor (ArgR), or redox-sensing transcriptional repressor (Rex). However, neither CodY, ArgR, nor Rex levels were changed following overexpression of SigS. As a result, any indirect SigS regulatory effects on these amino acid transport/metabolism genes are likely occurring through different transcription factors. Four small RNAs were also moderately increased (2- to 5-fold) following SigS induction, including SAOUHSCs125 (Ssr54), SAOUHSCs203, SAOUHSCs253, and SAOUHSCs215 (Fig. 1F and Table S5) (33, 34).
FIG 1.
The S. aureus SH1000 transcriptome following SigS overexpression. (A) Genetic constructs and experimental design for the SigS transcriptome experiment. Total RNA was isolated from S. aureus strains carrying either a plasmid-based xylose-inducible sigS construct or empty vector (EV) control grown in the presence of xylose to induce overexpression of SigS. (B) CIRCOS file demonstrating the S. aureus SH1000 transcriptome following overexpression of SigS. The genomic map depicts SH1000 EV (inner circle, black) and SH1000 pEPSA5-sigS (outer circle, red) transcriptomes, reported as transcripts per million (TPM) expression values. The outermost circle is a heatmap demonstrating fold change in expression, where red or blue indicates higher expression in the wild-type or sigS-overexpressing strain, respectively. (C) Heatmap of RNA sequencing data depicting S. aureus SH1000 transcripts whose expression was modulated at least 2-fold throughout the transcriptome following SigS induction. (D) Heatmap of SigS-modulated transcripts involved in immune evasion that were increased by at least 2-fold in reference to SigS and SroAB transcript levels. (E to G) As in panel D, but for small RNAs (E), the capsule biosynthesis operon (F), and SigB regulon members (G).
Seventy-five genes were downregulated in response to transient overexpression of SigS (Fig. 1B and C and Table S2). Many of the SigS-downregulated genes have no functional annotation (Fig. 1C and Table S2). Interestingly, five tRNA genes were repressed, including tRNA-Leu (trnaL), tRNA-Arg (trnaR), tRNA-Gly (trnaG), tRNA-Tyr (trnaY), and tRNA-Leu (trnaL) (Table S4). Thirteen of the SigS-repressed genes are part of the capsule biosynthesis operon (capABCDEFGHJKLMN [cap] operon) and are regulated by both SigB and CodY (Fig. 1E and Table S4). Fifty-three of the 75 SigS-downregulated genes are part of the SigB regulon (Fig. 1G; Tables S4 and S6). Finally, the expression of 10 previously discovered small RNAs was also decreased by at least 2-fold following SigS overexpression, SAOUHSCs47 (rsaI/rsaOG), SAOUHSCs042 (rsaOB), SAOUHSCs181 (Sau-6372), SAOUHSs221 (JKD6008sRNA178), SAOUHSs246 (JKD6008sRNA361), SAOUHSs274 (tsr24), SAOUHSCs065 (Sau-50), SAOUHSCs114 (ssr7), SAOUHSCs030 (sprB), and SAOUHSCs162 (teg40as) (Fig. 1F and Table S5). The transcription of SprB and SAOUHSCs162 (Teg40as) requires SigB (33, 35–40). To validate transcriptome sequencing (RNA-seq) findings, a selection of genes was assayed by real-time quantitative reverse transcription-PCR (RT-qPCR), with fold change in expression proving comparable to RNA-seq findings (Fig. S1).
The expression of a novel protein pair of unknown function is strongly activated by SigS.
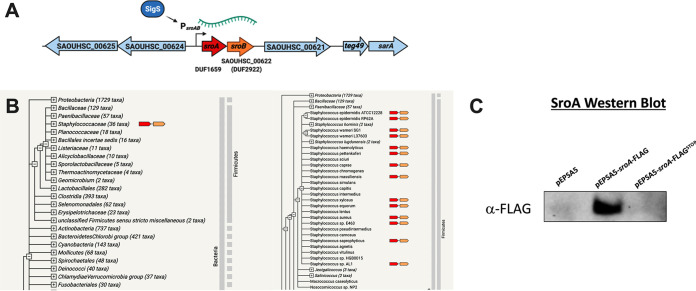
Beyond the changes noted above, the most highly upregulated genes that emerged from our SigS transcriptome experiment were two relatively short uncharacterized proteins, SACOL0676 (SAOUHSC_00622) and SACOL0677 (not annotated in the 8325 genome). These genes are encoded in tandem on a single polycistronic transcript and are increased 1,000-fold upon transient overexpression of SigS (Fig. 1C). Here, we rename them SigS-regulated OrfA (SroA) and SigS-regulated OrfB (SroB) (Fig. 2A and Fig. S2). The sroA gene encodes a 67-amino-acid protein with a DUF1659 domain (Fig. 2A and Fig. S2). The sroB gene encodes a 74-amino-acid protein with a DUF2922 domain (Fig. 2A and Fig. S2). There is a 16-bp region in between the stop codon of sroA and the start codon of sroB that contains a canonical Shine-Dalgarno sequence (AGGAGG) that is presumably the ribosome binding site for SroB translation (Fig. S2). There is also a canonical Shine-Dalgarno sequence (AGGAGG) starting 16 bp upstream of the predicted sroA start codon that is presumably the ribosome binding site for SroA translation. While the DUF1659 and DUF2922 domains are widely present in many bacterial species, there is little to no information about their function. Importantly, the amino acid sequences of the S. aureus SroA and SroB protein sequences are conserved only within the staphylococcal family. Computational analysis of SroA and SroB amino acid sequences using the web-based String database (https://string-db.org/) demonstrates strict co-occurrence within the staphylococcal family (Fig. 2B). The sroB gene, AureoWiki pan ID SAUPAN002490000, is annotated in 100% of the S. aureus strains in the AureoWiki metagenomic database. However, the sroA gene, AureoWiki pan ID SAUPAN002491000, is annotated in 88% of 33 strains within the AureoWiki metagenomic database (https://aureowiki.med.uni-greifswald.de/SAUPAN002491000). The sroA gene annotation is absent in the genome sequence maps of strains NCTC 8325, Newman, RF122, and TCH60. We therefore decided to confirm that the sroA gene encoded a protein in S. aureus SH1000. We constructed a xylose-inducible FLAG-tagged allele of sroA (sroA-FLAG). We also constructed a xylose-inducible FLAG-tagged allele of sroA with a STOP codon substitution mutation for the start codon (sroA-FLAGSTOP). We then performed Western blot analysis using an antibody to the FLAG tag on total protein isolated from xylose-induced exponentially growing cultures of SH1000 containing either (i) empty vector control, (ii) wild-type (WT) sroA-FLAG, or (iii) sroA-FLAGSTOP (Fig. 2C). We were able to detect SroA-FLAG protein levels from the wild-type sroA-FLAG allele. However, expression from the sroA-FLAGSTOP allele was undetectable, suggesting that the sroA gene does produce a protein (Fig. 2C).
FIG 2.
The sroAB genetic locus and co-occurrence within the Staphylococcaceae. (A) The sroAB genetic locus in S. aureus SH1000. Note that sroA is not annotated in the genome of this or other NCTC 8325 strains. (B) The amino acid sequence of SroA was translated from the S. aureus NCTC 8325 genome and entered into the String database (https://string-db.org/) to analyze for co-occurrence. (C) FLAG-tagged Western blotting of wild-type SroA or an allele with the start codon replaced with a stop codon (STOP). SroA was overexpressed in the log phase, growing S. aureus SH1000 from a plasmid-based xylose-inducible promoter.
SigS directly regulates the transcription of sroAB in response to DNA damage.
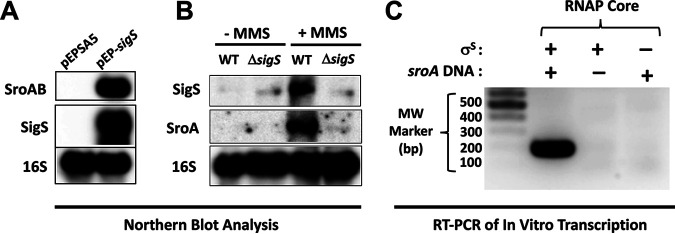
To confirm that SigS overexpression resulted in the upregulation of sroAB, we conducted sroAB Northern blot analysis on total RNA samples isolated from S. aureus following SigS overexpression (Fig. 3A). In the strain containing the vector control, both SigS and SroAB expression were completely undetectable (Fig. 3A). Conversely, in the strain containing the episomal xylose-inducible allele of SigS, SroA was activated along with SigS. We previously reported that SigS activity is stimulated following exposure to the DNA-damaging agent MMS (4, 21). We therefore reasoned that sroAB expression should also be stimulated in response to MMS treatment in a SigS-dependent manner if it is a regulatory target of SigS. To test this hypothesis, we grew wild-type and ΔsigS strains of S. aureus HOU (MRSA) in tryptic soy broth (TSB) supplemented with MMS and analyzed total RNA by Northern blotting analysis. Consistent with our previous results, the sigS transcript was increased in cultures treated with MMS (Fig. 3A). The sroA mRNA was also detected in cultures treated with MMS; however, in the absence of sigS, the sroA transcript was undetectable (Fig. 3B). This result further supports the idea that SroA is a regulatory target of SigS. To determine if SigS directly binds to the sroAB promoter, we performed in vitro transcription using RNA polymerase (RNAP) core enzyme complexed with purified recombinant SigS protein and analyzed transcriptional yields using endpoint RT-PCR (Fig. 3C). RNAP core reconstituted with purified recombinant SigS did indeed result in a transcription product, suggesting that the sroAB promoter region is recognized by the RNAP-σS complex and that sroAB is a direct target of SigS (Fig. 3C).
FIG 3.
Northern blotting and in vitro transcription of sroAB in response to SigS activity. (A) Overnight cultures of wild-type SH1000 carrying pEPSA5 or pEPSA5-sigS were subcultured (1:200) in 30 mL of TSB-Cm and grown to an OD600 of 0.3. Cultures were induced with xylose to a final concentration of 2% for 30 min, and total RNA was isolated. Total RNA (3 μg) was analyzed by Northern blotting using biotin-labeled probes for 16S rRNA, SigS mRNA, and SroA mRNA. (B) As in panel A, but using S. aureus HOU wild type and ΔsigS. Overnight cultures were subcultured (1:200) in 30 mL of TSB supplemented with MMS to a final concentration of 30 mM and grown to an OD600 of 0.3. (C) In vitro transcription of the sroAB promoter region using RNA polymerase core enzyme reconstituted with purified recombinant S. aureus SigS.
SroA and SroB have divergent effects on sigS mRNA and its antisense RNA transcript.
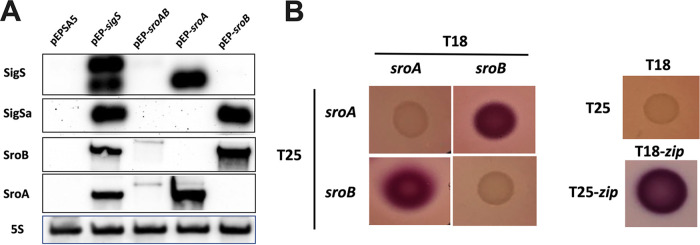
Due to the intense induction of the sroAB transcript in response to SigS overexpression, we hypothesized that SroAB may exhibit feedback regulation on sigS expression, as seen in other bacteria, as it pertains to regulatory circuits involving alternative sigma factors (25). Consequently, we overexpressed sroA, sroB, and sroAB and measured both the sigS mRNA and the previously described sigS cis-antisense RNA hypothesized to posttranscriptionally repress SigS production (22). This cis-antisense RNA is the 3′ untranslated region (UTR) of SAOUHSC_01899 (SACOL1829) and is annotated as s750 in the NCTC 8325 database. Remarkably, overexpression of SroA promotes sigS mRNA accumulation, while SroB overexpression promotes accumulation of s750 (sigS cis-antisense RNA) (Fig. 4A). Interestingly, overexpression of the complete sroAB transcript did not promote accumulation of either the sigS mRNA or its antisense transcripts (Fig. 4A).
FIG 4.
SigS and SigS antisense Northern blot analysis in the context of SroAB activity. (A) Overnight cultures of SH1000 carrying pEPSA5, pEP-sigS, pEP-sroAB, pEP-sroA, or pEP-sroB were subcultured in 30 mL of TSB-Cm and grown to an OD600 of 0.3. Cultures were induced with xylose to a final concentration of 2% for 30 min, and total RNA was isolated. Total RNA (3 μg) was analyzed by Northern blotting using biotin-labeled probes for 16S rRNA, SigS mRNA, SigSa (sigS antisense RNA resulting from the 3′ UTR of SAOUHSC_01899), SroA mRNA, and SroB mRNA. (B) Bacterial two-hybrid (BACTH) analysis of SroA and SroB. The sroA and sroB genes were cloned into BACTH plasmids pEB355 (T18) and pEB354 (T25) and analyzed on MacConkey-maltose agar plates at 30°C along with empty vector controls and positive controls pEB355-zip (T18-zip) and pEB354-zip (T25-zip).
We hypothesized that SroA and SroB may regulate each other through direct protein-protein interaction, effectively canceling out the activity of each other when they are both expressed. We decided to test this hypothesis by executing a bacterial two-hybrid assay (41). We spotted colonies cotransformed with plasmids encoding pEB355-sroA (T18-sroA) and pEB354-sroB (T25-sroA) or pEB355-sroB (T18-sroB) and pEB354-sroA (T25-sroA) in a 2 by 2 array, allowing us to test for potential SroA-SroB, SroA-SroA, and SroB-SroB interactions (Fig. 4B). Our positive and negative controls exhibited the expected Lac-positive (Lac+) and Lac-negative (Lac−) phenotypes, respectively (Fig. 4B). The SroA-SroA and SroB-SroB groups were both Lac− (Fig. 4B); however, the SroA-SroB spots both exhibited a Lac+ phenotype (Fig. 4B). Taken together, this suggests that SroA and SroB interact with each other while not interacting with themselves.
SroA inhibits SigS mRNA decay.
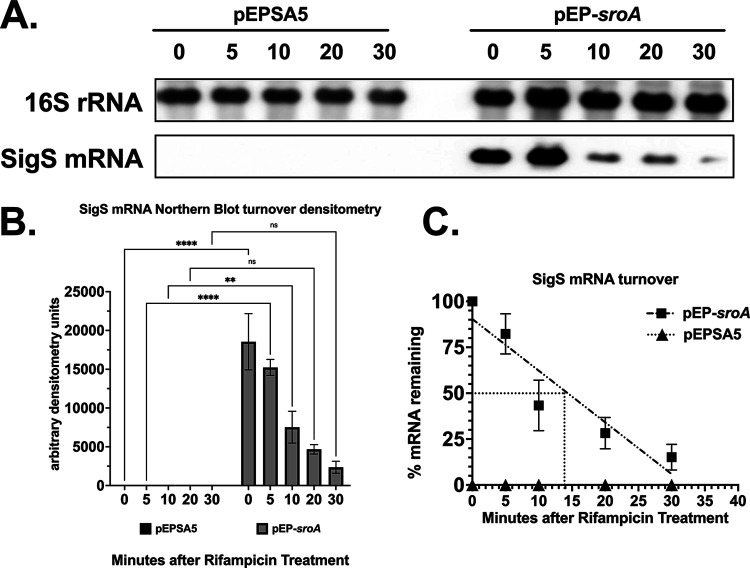
While our data suggested that SroA promotes accumulation of sigS mRNA levels in the absence of an interaction with SroB, it was not clear how SroA promotes SigS accumulation. Since SroA lacks a predicted DNA binding domain, we hypothesized that the SroA stimulatory effect on SigS was either through an indirect stimulatory effect on sigS transcription or a direct effect on sigS mRNA turnover. We thus decided to measure sigS mRNA decay following MMS induction using a rifampin chase assay (Fig. 5). The half-life (t1/2) of sigS mRNA from S. aureus SH1000 containing the vector control was less than 5 min, while the t1/2 of sigS mRNA isolated from S. aureus SH1000 overexpressing SroA was approximately 15 min. This suggests that SroA acts to inhibit sigS mRNA decay (Fig. 5).
FIG 5.
SigS mRNA turnover following overexpression of SroA. (A) Northern blot analysis of wild-type SH1000 carrying either pEPSA5 or pEP-sroA grown in 30 mL of TSB-Cm supplemented with MMS to a final concentration of 25 mM and an OD600 of 0.3. Cultures were then induced with xylose to a final concentration of 2% and incubated for an additional 30 min. Cells were harvested and resuspended in fresh TSB-Cm supplemented with rifampin to a final concentration of 500 μg/mL, and total RNA was isolated at 5- to 15-minute intervals. Total RNA (3 μg) was analyzed by Northern blotting using biotin-labeled probes for 16S rRNA and SigS mRNA. (B) Densitometry analysis of Northern blotting. (C) Calculation of the half-life (t1/2) of SigS mRNA based on densitometry analysis in panel B. Quantitative Northern blotting data from panel B were subjected to linear regression analysis using GraphPad Prism 9. Experiments were repeated at least three times, and data are presented as the mean plus or minus the standard error of the mean.
The sroAB operon is required for full virulence in female mice.
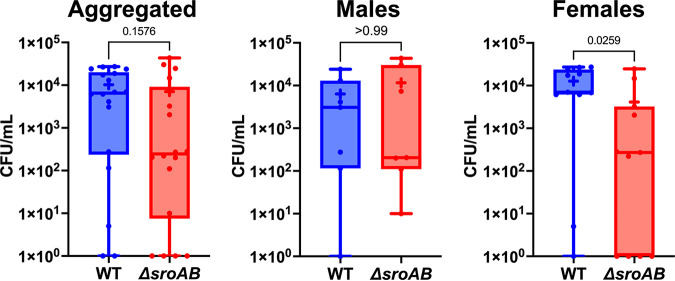
While SroA clearly acts to regulate SigS abundance at the posttranscriptional level, the role of the sroAB locus in virulence is undefined. We hypothesized that the sroAB locus is necessary for S. aureus virulence since SigS promotes virulence and SroA stimulates sigS expression. To test this hypothesis, we measured the virulence of a sroAB double mutant in a murine model of pneumonia. C57BL/6J mice were each infected intranasally with the wild type or ΔsroAB strains of S. aureus USA300 (Fig. 6). After 24 h, mice were sacrificed, lungs harvested, and bacterial burden determined as CFU per milliliter (Fig. 6). Upon analysis, we noted a statistically significant 3-fold decrease in the number of ΔsroAB mutants recovered from female mice compared to wild type (Fig. 6). Interestingly, there was no difference in the amount of wild-type or mutant cells recovered from male mice, for reasons that are not clear (Fig. 6).
FIG 6.
S. aureus virulence in a murine model of pneumonia in the absence of sroAB. C57BL/6J mice (7 male, 11 female) were each infected intranasally with 5 × 107 CFU of either USA300 (WT) or ΔsroAB. After 24 h, mice were sacrificed, lungs harvested, and bacterial burden determined. Shown are maximum and minimum values (whiskers) and 25th to 75th percentiles (boxes). Horizontal lines, means; +, medians. Statistical significance was determined using a Mann-Whitney test.
DISCUSSION
Through our transcriptome studies, we identified a previously uncharacterized locus encoding a bicistronic transcript that we named sroAB, which was increased 1,000-fold following SigS overexpression. The sroAB transcript was preeminent among the SigS-activated transcripts in our study. This suggests that SroA and SroB are major downstream effectors of the SigS stress response. SroA and SroB each encode midsized bacterial proteins with 68 and 72 amino acid residues, respectively. While these proteins are just above the upper limit of 50 amino acids for the definition of short ORFs in bacteria, their relatively small size is interesting to note, as investigations into the dark proteome and dark genome are increasing (42–45).
We also identified several immune evasion genes that were upregulated in response to SigS overexpression (Fig. 1C). Both Spa and Sbi are immunoglobulin binding proteins that would ultimately inhibit antibody opsonization to, phagocytosis of, and clearing of S. aureus infection. The other SigS-upregulated immune evasion genes, hlgBC and lukAB, encode leukotoxins that are effective in the targeting of various leukocytes. Both operons are also stimulated by the SaeRS two-component signal transduction system that senses human neutrophil peptides (46). SigS-dependent upregulation of sbi, spa, and hlgBC may provide a mechanism to explain the increased sensitivity of ΔsigS mutants to whole human blood and macrophages (21).
Our observation of SigS-dependent upregulation of amino acid transport/metabolism genes is congruent with prior analysis of SigS regulation (23). Burda et al. isolated several transposon mutants that resulted in either activation or repression of sigS expression, some of which were localized to genes involved in amino acid transport or metabolism (23). These include mutants in two apparently cistronic genes that are both strongly upregulated in response to SigS overexpression in our study, ald1 (SAOUHSC_01452/SACOL1478) and SAOUHSC01450 (SACOL1476) (see Tables S2 and S7 in the supplemental material). The remaining gene in that operon, ilvA1, is also upregulated in response to SigS overexpression (Tables S2 and S7). Interestingly, based on the results of the transposon mutant screen, ald1 and SAOUHSC01450 (SACOL1476) have opposing regulatory effects on sigS expression for reasons that are not clear (Table S7). We also identified four SigS-stimulated targets involved in arginine or glutamine metabolism. Taken together, our transcriptome results support the model, previously established by Burda et al. (23), that SigS is involved in the sensing and regulation of amino acid transport and metabolism (Table S7). Additionally, many of the SigS-repressed genes are a part of the CodY regulon involved in BCAA/GTP sensing, mainly the capsule biosynthesis genes (Fig. 1E and Table S4). The capsule biosynthesis and many of the other SigS-repressed genes are a part of the SigB regulon. Repression of SigB-activated genes upon overexpression of SigS could be due to sigma factor competition (SigS versus SigB) for core RNA polymerase (47, 48). Yet the influence of sigma factor competition in Firmicutes is not clear.
A major function that we uncovered was the ability of SroA to promote positive feedback regulation of SigS. Previous studies have demonstrated that in vitro expression of sigS is very low. Several chemicals stimulate transcription of sigS, most notably the chemical mutagen MMS (4, 21). Following MMS treatment, sigS transcription is strongly increased. However, the mechanisms used to tightly control sigS expression and transduce chemical stressors to stimulate gene expression are still unclear. Biochemical and genetic screens for sigS transcriptional regulators yielded several candidates, some of which bind to the sigS promoters, including KdpE and CymR (4, 21, 22), while ArlR and LacR are indirect regulators of sigS transcription (4, 21, 22). Tight regulation of sigS transcription is likely to be part of a mechanism used by S. aureus to repress SigS levels and conserve cellular resources until SigS is needed for restoration of homeostasis after exposure to various stressors. However, transcriptional control is only likely to be part of a collection of regulatory switches needed to tightly control SigS levels.
Posttranscriptional control of gene expression usually includes small RNA regulation of translation or mRNA turnover. Prior to this work, little was known about the posttranscriptional regulatory control of SigS. Interestingly, we demonstrate a role for the sroAB locus in posttranscriptional regulatory control of SigS. We identified a role for SroA in the stabilization of the SigS mRNA transcript. SroA stabilization of the SigS mRNA is particularly interesting given the intrinsically low levels of SigS and incomplete picture of SigS regulatory control. In addition to tight transcriptional control, low levels of endogenous SigS expression could be due to SigS mRNA instability, and SroA activity could mitigate this instability to transiently increase SigS levels. Interestingly, overexpression of SroB, but not SroAB together, did not increase SigS levels. A positive bacterial two-hybrid interaction between SroA and SroB suggests that an SroA-SroB interaction inhibits the ability of SroA to stabilize SigS mRNA.
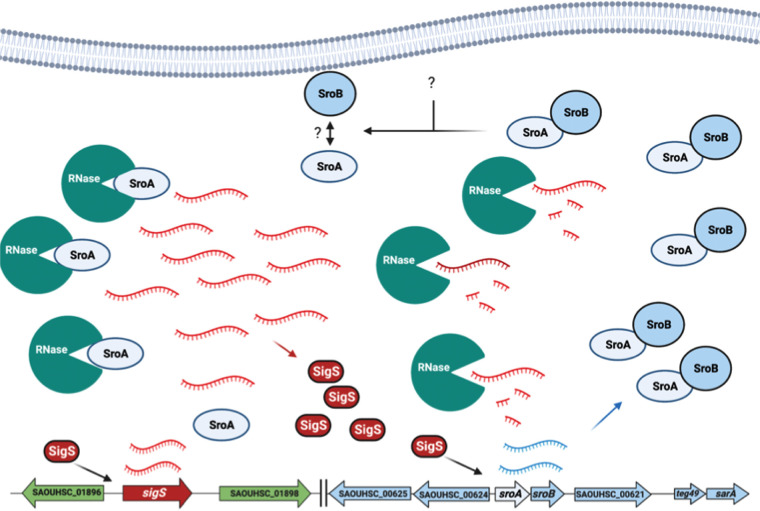
Based on the data articulated above, we have created a model for a SigS-SroAB regulatory circuit in S. aureus (Fig. 7). In the presence of DNA damage, sigS transcription is increased. However, the level of SigS protein that is synthesized is relatively low due to high mRNA turnover. The SigS protein that is synthesized directly stimulates transcription of the sroAB locus, leading to the expression of SroA and SroB. While SroA can stimulate SigS accumulation at the level of mRNA stability, a direct interaction of SroA and SroB inhibits this. Under a condition that has yet to be identified, the SroA-SroB interaction is inhibited. This inhibition could occur through posttranslational modification of SroA or SroB. Alternatively, SroB may be degraded by an intracellular protease. In either case, the resulting free SroA inhibits ribonucleolytic activity targeting SigS. However, the precise role of RNases in SigS mRNA turnover and the molecular mechanisms that control the SroA-SroB interaction are not yet clear. These phenomena are the subject of ongoing investigations in our laboratory.
FIG 7.
Model for the SigS-SroAB regulatory circuit. In response to the DNA-damaging agent MMS, SigS is activated (through an unknown mechanism). SigS acts to stimulate sroAB expression. SroA and SroB interact with each other under normal circumstances. In response to an unidentified internal or external signal, SroA-SroB association is inhibited through a mechanism that is unclear. Then, free SroA promotes SigS accumulation through inhibition of RNases that promote SigS transcript degradation.
Prior to this work, it was well established that SigS was necessary for S. aureus survival in the presence of chemical stressors, environmental stressors, and exposure to immune cells, including ethanol, H2O2, cell wall-targeting antibiotics, chemical mutagens (MMS), elevated temperature, and intracellular macrophage survival. We aimed to identify downstream effectors of the SigS stress response to further understand its function in S. aureus infection. Our work supports a role for the sroAB locus in S. aureus virulence and the need for further studies of SroAB function, which will increase our understanding of S. aureus virulence and SigS-mediated pathogenesis, immune evasion, and stress response.
MATERIALS AND METHODS
Media and growth conditions.
All S. aureus strains were grown in tryptic soy broth (TSB). Overnight cultures of S. aureus were grown in 5 mL of TSB within a roller drum placed inside a microbiological incubator at 28°C or 37°C. S. aureus strains containing plasmids were grown in TSB supplemented with chloramphenicol or erythromycin to a final concentration of 10 μg/mL (TSB-Cm or TSB-Erm). S. aureus RN4220 plasmid transformants were selected on TSB-Cm agar or TSB-Erm agar plates (TSA-Cm or TSA-Erm). Phage 80α-mediated plasmid or mutant transductants were selected on TSA-Cm or TSA-Erm plates supplemented with sodium citrate (NaCi) to a final concentration of 5 mM (TSA-Cm-NaCi or TSA-Erm-NaCi). E. coli strains used for propagation of recombinant DNA were grown in Luria-Bertani-Lennox broth (LB), or LB agar plates, supplemented with ampicillin to a final concentration of 100 μg/mL (LB-Amp), kanamycin to a final concentration of 25 μg/mL (LB-Kan), or chloramphenicol to a final concentration of 10 μg/mL or 25 μg/mL (LB-Cm). E. coli cotransformants for BACTH analysis were selected on LB agar plates supplemented with ampicillin to a final concentration of 100 μg/mL and kanamycin to a final concentration of 25 μg/mL (LB-Amp-Kan). Overnight cultures of E. coli were grown in 5 mL of LB within a roller drum placed inside a microbiological incubator at 30°C or 37°C.
Strains, plasmids, and oligonucleotides.
All bacterial strains used in this study are listed in Table 1. All plasmids used in this study are listed in Table 2. Oligonucleotides used for PCR amplification, one-step reverse transcriptase PCR (RT-PCR), real-time quantitative reverse transcription-PCR (RT-qPCR) analysis, and as biotinylated Northern blot probes are listed in Table S1 in the supplemental material.
TABLE 1.
List of strains used in this study
| Strain name | Bacteria or genotype | Source and/or comments |
|---|---|---|
| E. coli strains | ||
| BTH101 | F−, cya-99, araD139, galE15, galK16, rpsL1 (Strr), hsdR2, mcrA1, mcrB1 | Aureilia Battesti |
| NEB5α | fhuA2 Δ(argF-lacZ) U169 phoA glnV44 Φ80Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | New England Biolabs |
| KMTE-167 | NEB5α pEPSA5 (Ampr) | NEB5α + pEPSA5, heat shock; this work |
| KMTE-168 | NEB5α pEP-sigS (Ampr) | NEB5α + pEP-sigS, heat shock; this work |
| KMTE-169 | NEB5α pEP-sroAB (Ampr) | NEB5α + pEP-sroAB, heat shock; this work |
| KMTE-170 | NEB5α pEP-sroA (Ampr) | NEB5α + pEP-sroA, heat shock; this work |
| KMTE-171 | NEB5α pEP-sroA-FLAG (Ampr) | NEB5α + pEP-sroA-FLAG, heat shock; this work |
| KMTE-179 | NEB5α pIMAY* (Cmr) | NEB5α + pIMAY*, heat shock; this work |
| KMTE-190 | NEB5α pEP-sroA-FLAGSTOP (Ampr) | NEB5α + pEP-sroA-FLAGSTOP, heat shock; this work |
| KMTE-233 | NEB5α pIMAY*-ΔsroAB::ermC (Cmr) | NEB5α + pIMAY*-ΔsroAB::ermC, heat shock; this work |
| KMTE-263 | NEB5α pEP-sroB (Ampr) | NEB5α + pEP-sroB, heat shock; this work |
| S. aureus strains | ||
| RN4220 | Restriction minus derivative of 8325-4 laboratory strain 8325-4 r− m+ | Lab collection |
| KMTS-229 | RN4220 pEPSA5 (Cmr) | RN4220 + pEPSA5, electroporation; this work |
| KMTS-230 | RN4220 pEP-sigS (Cmr) | RN4220 + pEP-sigS, electroporation; this work |
| KMTS-231 | RN4220 pEP-sroAB (Cmr) | RN4220 + pEP-sroAB, electroporation; this work |
| KMTS-232 | RN4220 pEP-sroB (Cmr) | RN4220 + pEP-sroB, electroporation; this work |
| KMTS-233 | RN4220 pEP-sroA (Cmr) | RN4220 + pEP-sroA, electroporation; this work |
| KMTS-234 | RN4220 pEP-sroA-FLAG (Cmr) | RN4220 + pEP-sroA-FLAG, electroporation; this work |
| KMTS-234b | RN4220 pEP-sroA-FLAGSTOP (Cmr) | RN4220 + pEP-sroA-FLAGSTOP, electroporation; this work |
| KMTS-304 | RN4220 pIMAY*-ΔsroAB::ermC (Cmr) | RN4220 + pIMAY*-ΔsroAB::ermC, electroporation; this work |
| 8325-4 | Wild-type laboratory strain: 8325-4 rsbU− | Lab collection |
| SH1000 | Wild-type laboratory strain, 8325-4 rsbU+ | Lab collection |
| KMTS-216 | SH1000 pEPSA5 (Cmr) | SH1000 × Φ80α (KMTS-229: pEPSA5) |
| KMTS-217 | SH1000 pEP-sigS (Cmr) | SH1000 × Φ80α (KMTS-230: pEPSA5-sigS) |
| KMTS-218 | SH1000 pEP-sroAB (Cmr) | SH1000 × Φ80α (KMTS-231: pEPSA5-sroAB) |
| KMTS-219 | SH1000 pEP-sroB (Cmr) | SH1000 × Φ80α (KMTS-232: pEPSA5-sroB) |
| KMTS-220 | SH1000 pEP-sroA (Cmr) | SH1000 × Φ80α (KMTS-233: pEPSA5-sroA) |
| KMTS-248 | SH1000 ΔsroAB::erm (Ermr), sroAB− | SH1000 pIMAY*-ΔsroAB::ermC, following mutagenesis and plasmid excision |
| KMTS-250 | SH1000 pEP-sroA-FLAG (Cmr) | SH1000 × Φ80α (KMTS-234: pEPSA5-sroA-FLAG) |
| KMTS-251 | SH1000 pEP-sroA-FLAGSTOP (Cmr) | SH1000 × Φ80α (pEPSA5-sroA-FLAGSTOP) |
| KMTS-264 | SH1000 ΔsigS::tet (Tetr), sigS− | SH1000 × Φ80α (KMTS-242 - USA300 HOU ΔsigS::tet) |
| KMTS-241 | USA300 HOU | Lab collection, Lindsey Shaw |
| KMTS-242 | USA300 HOU ΔsigS::tet (Tetr), sigS- | Lab collection, Lindsey Shaw |
TABLE 2.
Plasmid list
| Plasmid | Characteristics | Source |
|---|---|---|
| pEPSA5 | Shuttle vector p15A ori in E. coli, bla (Ampr) in E. coli, cat (Cmr) in S. aureus | Prahathees Eswara |
| pEP-sigS | sigS gene cloned into the EcoRI/BamHI site of pEPSA5, bla (Ampr) in E. coli, cat (Cmr) in S. aureus | This work |
| pEP-sroA | sroA gene cloned into the EcoRI/BamHI site of pEPSA5, bla (Ampr) in E. coli, cat (Cmr) in S. aureus | This work |
| pEP-sroA-FLAG | sroA-FLAG gene cloned into the EcoRI/BamHI site of pEPSA5, bla (Ampr) in E. coli, cat (Cmr) in S. aureus | This work |
| pEP-sroA-FLAGSTOP | TGA-ATG start codon substitution in sroA-FLAG gene cloned into the EcoRI/PstI site of pEPSA5; bla (Ampr) in E. coli, cat (Cmr) in S. aureus | This work |
| pEP-sroB | sroB gene cloned into the EcoRI/BamHI site of pEPSA5, bla (Ampr) in E. coli, cat (Cmr) in S. aureus | This work |
| pEP-sroAB | sroAB gene cloned into the EcoRI/BamHI site of pEPSA5; bla (Ampr) in E. coli, cat (Cmr) in S. aureus | This work |
| pEB354 | Bacterial two-hybrid interaction expression vector, PLac, LacO site, CAP binding site, p15A ori, Cyaa T25 domain linker (Kanr) | Aureilia Battesti |
| pEB354-zip | zip domain gene cloned into the EcoRI/XhoI site of Cya in T25 domain linker of pEB354 (Kanr), T25-zip | Aureilia Battesti |
| pEB354-sroA | sroA domain gene cloned into the EcoRI/XhoI site of Cya in T25 domain linker of pEB354 (Kanr), T25-sroA | This work |
| pEB354-sroB | sroB domain gene cloned into the EcoRI/XhoI site of Cya in the T25 domain linker of pEB354 (Kanr), T25-sroB | This work |
| pEB355 | Bacterial two-hybrid interaction expression vector, PLac, LacO site, CAP binding site, ColE1 ori, Cya T18 domain linker, bla (Ampr) | Aureilia Battesti |
| pEB355-zip | zip domain gene cloned into the EcoRI/XhoI site of Cya in T18 domain linker of pEB355, bla (Ampr), T18-zip | Aureilia Battesti |
| pEB355-sroA | sroA domain gene cloned into the EcoRI/XhoI site of Cya in T18 domain linker of pEB355, PLac, bla (Ampr), “T18-sroA” | This work |
| pEB355-sroB | sroB domain gene cloned into the EcoRI/XhoI site of Cya in T18 domain linker of pEB355, PLac, bla (Ampr), T18-sroB | This work |
| pIMAY* | Allelic exchange plasmid, PheS* counterselection, cat (Cmr) in S. aureus and E. coli, temp-sensitive ori (orits) in S. aureus | pIMAY* was a gift from Angelika Grundling (Addgene plasmid number 121441; https://www.addgene.org/121441/; RRID Addgene_121441) |
| pIMAY*-ΔsroAB::ermC | ΔsroAB::ermC ± 1,000 bp PCR product cloned into EcoRI/XhoI site, cat (Cmr) | This work |
Cya, adenylate cylase.
(i) S. aureus strain engineering. S. aureus strains used in this study were derivatives of NCTC 8325 or the USA300 pulsed-field gel electrophoresis (PFGE) type (Table 1) (49). S. aureus strains were genetically engineered using electroporation and transduction. E. coli-S. aureus plasmid shuttle vectors were electroporated into S. aureus restriction-minus strain RN4220, as previously described (50, 51). Phage 80α lysates were then created on S. aureus RN4220 strains carrying plasmids, on TSA-Cm or TSA-Erm, supplemented with CaCl2 to a final concentration of 5 mM (TSA-Cm-CaCl2 or TSA-Erm-CaCl2), using the top agar method as previously described (52). Plasmid constructs then were moved from RN4220 into SH1000 by ϕ80α transduction as previously described (53). The ΔsigS::tet mutant was also moved from the USA300 PFGE-type strain background to the SH1000 strain background using phage 80α transduction as previously described (53). The chromosomal ΔsroAB::ermC mutation was constructed in strain SH1000 using pIMAY* after cloning a ΔsroAB::ermC into pIMAY* as previously described (54).
(ii) E. coli strain engineering. E. coli strains used in this study were for the purposes of cloning and bacterial adenylate cyclase two-hybrid (BACTH) analysis and are listed in Table 1. All molecular cloning was executed in E. coli NEB5α (New England Biolabs) (Table 1). All BACTH analysis was executed in an adenylate cyclase mutant (cya−) strain of E. coli BTH101 (41, 55).
(iii) Plasmid backbones used for expression, mutagenesis, and BACTH analysis. Plasmids used in this study are listed in Table 2. Plasmid expression vectors are derivatives of pEPSA5, an E. coli-S. aureus shuttle vector containing a xylose-inducible promoter and its cognate regulator, XylR (Table 2) (56). The sroAB mutagenesis vector is a derivative of pIMAY*, a temperature-sensitive plasmid using pheS* as a counterselectable marker. Plasmids used for BACTH analysis are derivatives of pEB355 (T18) or pEB354 (T25) (41, 55). Construction of the plasmid derivatives is described below. All plasmids containing inserts or site-directed point mutations were identified by colony PCR, verified by PCR amplification, and confirmed by Sanger sequencing. All resulting recombinant DNA constructs were transformed into NEB5α or NEB10β (New England Biolabs) using heat shock transformation, according to the manufacturer’s instructions.
(iv) Construction of pEPSA5 derivatives. For these studies, we created several plasmid-based xylose-inducible constructs via standard recombinant DNA technology techniques, cloning desired genes for expression into pEPSA5. We created pEPSA5 derivatives containing sigS, sroA, sroB, sroAB, and sroA-FLAG. Briefly, we amplified the respective genes from SH1000 genomic DNA using oligonucleotide primers listed in Table S1. Both purified pEPSA5 and the PCR products to be inserted were digested with EcoRI and BamHI and ligated using Instant Sticky-End ligase mastermix (New England Biolabs). To construct a nonsense mutation in the sroA start codon within the pEP-sroA-FLAG construct (to create pEP-sroA-FLAGSTOP), we used mutagenic primers KT1604 and KT1606 and the Q5 site-directed mutagenesis kit (New England Biolabs) according to the manufacturer’s instructions.
(v) Construction of pIMAY*-sroAB::ermC for the sroAB mutagenesis plasmid. We constructed recombinant plasmids for the creation of erythromycin-marked deletion-insertion mutations of sroAB (pIMAY*-ΔsroAB::ermC). We created pIMAY*-ΔsroAB::ermC using HiFi DNA assembly. First, we amplified three different DNA sequences, (i) “ΔsroAB::ermC up” from S. aureus SH1000 genomic DNA, using primers KT1757 and KT1758, (ii) “ΔsroAB::ermC internal” from S. aureus shuttle vector pCN51, using primers KT1659 and KT1660, and (iii) “ΔsroAB::ermC down” from S. aureus SH1000 genomic DNA, using primers KT1759 and KT1760. The “ΔsroAB::ermC up” PCR product has approximately 30 bp of homology to the region upstream of the multiple-cloning site (MCS) of pIMAY* on its 5′ end and approximately 30 bp of homology to the 5′ end of the “ΔsroAB::ermC internal” PCR product on its 3′ end. The “ΔsroAB::ermC down” PCR product has approximately 30 bp of homology to the region downstream of the MCS of pIMAY* on its 3′ end and approximately 30 bp of homology to the 3′ end of the “ΔsroAB::ermC internal” PCR product on its 5′ end. We then digested pIMAY* with both EcoRI and XhoI. The resulting pIMAY* EcoRI/XhoI digest and the three PCR products were joined using the NEBuilder HiFi DNA assembly mastermix (New England Biolabs).
(vi) Construction of plasmids for BACTH analysis. We created recombinant plasmids containing adenylate cyclase cloned in-frame with sroA or sroB in BACTH vectors pEB355 (T18) and pEB354 (T25) (41, 55). The sroA and sroB genes were amplified by PCR, using their corresponding primers listed in Table S1, and digested with EcoRI and XhoI. We then ligated the purified sroA or sroB with purified pEB355 (T18) or pEB354 (T25) EcoRI/XhoI digests using Instant Sticky-End ligase and transformed them into chemically competent NEB5α cells according to the manufacturer’s instructions. Transformants were selected on LB-Amp or LB-Kan plates.
Total RNA isolation.
Total RNA was isolated using the Fast RNA Pro Blue kit (MP Biomedicals). Briefly, S. aureus cells of interest were harvested from 5 to 50 mL of culture at the selected condition (optical density at 600 nm [OD600] and/or specific experimental treatment). Pellets were resuspended in 1 mL of RNAPro solution, added to silica beads, and homogenized using the Precellys 24 dual homogenizer (Bertin Corporation) with the Cryolys for Precellys adaptor to ensure integrity of RNA following heat exposure. Following lysis, RNA extraction was continued via chloroform extraction and ethanol precipitation overnight at −80°C. RNA cell pellets were resuspended in 50 to 100 μL of RNase-free H2O. RNA concentration was measured using a NanoDrop.
RNA sequencing.
RNA sequencing and data analysis were performed as described previously (57). Briefly, the SH1000 wild type carrying either empty pEPSA5 or pEP-sigS was grown overnight as detailed above. Next, these cultures were used to inoculate fresh TSB and allowed to grow for a further 3 h. After this time, these cultures were used to seed fresh TSB at an OD600 of 0.05. Strains were allowed to grow until exponential phase (OD600 = 0.3) before the addition of xylose to a final concentration of 2% to induce expression of sigS. These cultures were grown for 30 min before samples were combined with 5 mL ice-cold phosphate-buffered saline (PBS) and subject to centrifugation. Total RNA extractions were performed using a Qiagen RNeasy kit, and DNA was removed with the Ambion Turbo DNA-free kit. RNA quality was determined using an Agilent 2100 Bioanalyzer with an RNA 6000 Nano kit to confirm RNA integrity (RIN). Only samples with a RIN of 9.7 or higher were used in this study. Triplicate samples for each strain, from independently grown cultures, were then pooled at equal RNA concentrations, followed by rRNA removal using a MicrobExpress bacterial mRNA enrichment kit. Efficiency of rRNA removal was confirmed using an Agilent 2100 Bioanalyzer with an RNA 6000 Nano kit. These mRNA samples were then subject to library preparation using a TruSeq stranded mRNA kit (Illumina) with the mRNA enrichment steps omitted. Fragment size, quantity, and quality were assessed using an Agilent 2100 Bioanalyzer with an RNA 6000 Nano kit. Library concentrations for the pooling of barcoded samples were assessed with a KAPA library quantification kit. Samples were run on an Illumina NextSeq with a 150-cycle NextSeq mid-output kit v2.5. Data were exported from BaseSpace (Illumina) in fastq format and uploaded to CLC Genomics Workbench for analysis. Data were aligned to the NCTC 8325 reference genome file, reannotated to include newly discovered small RNAs (GenBank accession no. NC_007795.1) (33). Comparisons were carried out following quantile normalization via the Qiagen Bioinformatics experimental fold change feature.
RT-qPCR transcriptional analysis.
To validate RNA-seq findings, a selection of genes was assayed by RT-qPCR. Strains were grown, RNA harvested, DNA removed, and sample quality assessed as described above. One microgram from each sample was reverse transcribed using an iScript cDNA synthesis kit (Bio-Rad). RT-qPCR was then performed using gene-specific primers (Table S1) and TB green premix Ex Taq (TaKaRa). Levels of gene expression were normalized to the 16S rRNA gene, and fold change of expression was assessed for sigS overexpression relative to wild-type samples, using the threshold cycle (2−ΔΔCT) method.
In vitro transcription of sroA with RNAP-σS.
One unit of RNA polymerase core enzyme (New England Biolabs) was added to 1 μg of purified recombinant σS (Biomatik) in 1 μL of 5× RNA polymerase reaction buffer (New England Biolabs) and incubated at 4°C for 15 min. After incubation, 1 μg of purified PCR product corresponding to the sroA coding region and 980 bp of upstream DNA was added to the σS-core-RNA polymerase mixture and incubated at 37°C for 15 min. Then, ribonucleoside triphosphates (rNTPs) were added to initiate transcription, and the reaction mixture was incubated at 37°C for 30 min. After this time, RNA was purified twice by acid phenol-chloroform extraction followed by ethanol precipitation. RNA was also purified by the Turbo DNA-free kit (Thermo Scientific) following the manufacturer’s protocol. A one-step RT-PCR was then executed on the purified RNA using primers KT1578/KT1579 (creating a product corresponding to the start codon of sroA to the termination codon of sroA). This experiment was repeated with two controls, purified σS or core-RNA polymerase. RT-PCRs were then run on a 2% agarose gel and imaged using a FluorChem R system (Protein Simple).
BACTH analysis.
We analyzed protein-protein interactions in vivo using the previously described BACTH system (41, 55). Briefly, we coelectroporated either (i) putative interaction pairs pEB355-sroA (T18-sroA) and pEB354-sroB (T25-sroB), (ii) putative interaction pairs pEB355-sroB (T18-sroB) and pEB354-sroA (T25-sroA), (iii) empty vector control pairs pEB355 (T18) and pEB354 (T25), or (iv) positive-control pairs pEB355-zip (T18-zip) and pEB354-zip (T25-zip) into E. coli BTH101. We allowed cells to recover at 30°C for several hours and plated them on LB agar plates supplemented with ampicillin and kanamycin. Then, several cotransformants were inoculated into 5 mL of LB-amp-kan media supplemented with IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 100 μM and incubated overnight at 30°C. A 10-μL aliquot of overnight cultures was spotted onto MacConkey-Maltose plates supplemented with IPTG to a final concentration of 100 mM and incubated at 30°C for 24 to 48 h to measure the Lac phenotype.
SigS rifampin chase assay.
SH1000 containing either pEPSA5 (empty vector control) or pEP-sroA was grown in TSB-Cm supplemented with MMS to a final concentration of 25 mM at 37°C to an OD600 of 0.5. Cells were harvested to remove supplemental MMS and resuspended in fresh TSB-Cm supplemented with xylose to a final concentration of 2% before being incubated for an additional 30 min. Cells were then harvested again by centrifugation to remove supplemental xylose and resuspended in fresh TSB-Cm. Rifampin was added to a final concentration of 500 mM to stop intracellular transcription, and total RNA was isolated at 2- to 5-min intervals following rifampin supplementation. We then subjected these samples to Northern blot analysis.
Northern Blot analysis.
A 1% MOPS (morpholinepropanesulfonic acid) agarose gel was used for resolution of total RNA. The agarose gel and buffer were created using UltraPure agarose (Invitrogen) in 1× MOPS buffer, diluted 1:10 from 10× MOPS buffer (Quality Biological, Inc.) mixed with diethyl pyrocarbonate (DEPC) water and 2 μL EtBr (10 μg/mL). The gel was prerun at 100 V for 40 minutes. Samples were mixed with urea gel loading buffer (National Diagnostics), heated at 65°C for 15 minutes, and resolved on the gel. The gel was soaked in 0.05 M NaOH solution for 20 minutes and then upside down in 20× SSC “(1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)” solution for 1 h. Samples were transferred to membrane using the capillary method and cross-linked with short-wave UV light to fix the transferred RNA. The cross-linked membrane was then prehybridized with 5 mL of PerfectHyb Plus hybridization buffer (MilliporeSigma) for 2 h. Following this, a biotinylated DNA probe corresponding to either SigS or 16S rRNA was added to the hybridization buffer to a final concentration of 500 ng/mL and hybridized at 42°C for 2 h. The membranes were then processed using stringency washes and developed using a chemiluminescent nucleic acid detection module kit (Thermo Fisher Scientific) according to the manufacturer’s recommendations. The chemiluminescent signal was detected using FluorChem R. Northern blot densitometry signals were quantified using ImageJ and analyzed using GraphPad Prism.
Total protein isolation.
To isolate total protein, S. aureus cells were harvested from 10-mL culture aliquots. Cells were pelleted and resuspended in 1 mL of 1× PBS and processed twice in the Precellys dual homogenizer for 40 s at a setting of 5,000 rpm before being centrifuged at 15,000 × g for 5 min at 4°C. Proteins were precipitated from the solution via treatment with trichloroacetic acid (TCA) at a final concentration of 25% on ice followed by centrifugation at 15,000 rpm for 5 min at 4°C. The pellet was washed using 200 μL of 100% ice-cold acetone (Thermo Scientific) before being resuspended in 50 μL of 1× PBS. Protein concentration was determined using the detergent-compatible (DC) protein assay kit (Bio-Rad).
SDS-PAGE and Western blot analysis.
Total protein (30 μg) was mixed with 4× NuPAGE LDS sample buffer (Thermo Scientific) and NuPAGE sample reducing agent (Thermo Scientific) before being heated at 70°C for 10 min and then cooled on ice for 2 min. The samples were then loaded onto a 12% Bis-Tris miniprotein gel (Thermo Scientific) and subjected to electrophoresis using 1× MES (morpholineethanesulfonic acid) buffer (Thermo Scientific) at 100 V for 1 h. The protein gel was transferred to a 0.45-μm-pore-size nitrocellulose membrane using the Trans-Blot Turbo system (Bio-Rad) according to the manufacturer’s instructions. The membrane was washed for 5 min three times with PBS with Tween 20 (PBS-T) buffer. Then, the membrane was washed in blocking solution (5% blotting-grade nonfat milk in 1× PBS-T buffer) for 30 to 120 min at room temperature with gentle agitation. The membrane was then washed three times with 1× PBS-T with gentle agitation. Next, the membrane was incubated in blocking solution containing the primary antibody to the FLAG epitope tag, α-FLAG (Sigma-Adrich), diluted 1:10,000 overnight. Following three washes with PBS-T for 5 min, the membrane was incubated for 2 h in blocking solution containing the secondary antibody diluted 1:25,000. The membrane was then washed three times with PBS-T for 5 min. The membrane was then incubated with chemiluminescent detection solution (Thermo Scientific), prepared by mixing 1 mL of chemiluminescent substrate and 50 μL of substrate enhancer for 5 min. The signal was developed using the FluorChem R system.
Mouse pneumonia model of infection.
Male and female 6-week-old C57BL/6J mice were purchased from Charles River Laboratories and allowed to acclimate for 1 week prior to infection. To prepare inocula, cultures of the wild-type and mutant strains were grown overnight (37°C, 250 rpm) before being diluted 1:100 in fresh TSB. After this time, strains were grown for 3 h before being standardized to an OD600 of 0.05. These cultures were then allowed to grow for 15 h before the CFU per milliliter were determined. This process was repeated three separate times, and the average CFU per milliliter was used to calculate the volume of bacteria required to generate a 10-mL inoculum of 1 × 108 CFU/30 μL. On the day of infection, cultures were grown as described, and the calculated volume of bacteria was harvested by centrifugation and washed with PBS before being resuspended in 10 mL PBS. Mice were anesthetized with isoflurane and then inoculated intranasally with 30 μL of the prepared bacteria suspension. Infections were monitored for 24 h or until mice reached a premoribund state, at which point they were euthanized. Lungs were harvested and homogenized in 2 mL PBS, and the bacterial burden (CFU/mL) was determined by serial dilution and plating. A Mann-Whitney test was used to determine statistical significance between the mutant and wild-type-infected mice.
Statistical analysis.
All statistical analysis was executed using Prism 9 software (GraphPad).
Ethics statement.
The mouse infection experiments were performed with the prior approval of the University of South Florida Institutional Animal Care and Use Committee.
Data availability.
Experimental data from this study were deposited in the NCBI Gene Expression Omnibus (GEO) database (GEO accession number GSE215075).
ACKNOWLEDGMENTS
This study was supported (in part) by several funding sources. These include a grant from the National Institute on Minority Health and Health Disparities of the National Institutes of Health under award number 2U54MD007597, grants AI124458 and AI157506 (L.N.S.) from the National Institutes of Allergy and Infectious Diseases, a grant from The Bridge Fund and Pilot Project Program (K.M.T.) of Howard University College of Medicine, and, finally, a grant from The Founders Chair in Basic Science from The Howard University Medical Alumni Association (K.M.T.).
We thank members of the Thompson and Shaw labs for critical review of the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Karl M. Thompson, Email: karl.thompson@Howard.edu.
George O’Toole, Geisel School of Medicine at Dartmouth.
REFERENCES
- 1.Cheung GYC, Bae JS, Otto M. 2021. Pathogenicity and virulence of Staphylococcus aureus. Virulence 12:547–569. doi: 10.1080/21505594.2021.1878688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fagerlund A, Granum PE, Havarstein LS. 2014. Staphylococcus aureus competence genes: mapping of the SigH, ComK1 and ComK2 regulons by transcriptome sequencing. Mol Microbiol 94:557–579. doi: 10.1111/mmi.12767. [DOI] [PubMed] [Google Scholar]
- 3.Horsburgh MJ, Aish JL, White IJ, Shaw L, Lithgow JK, Foster SJ. 2002. SigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J Bacteriol 184:5457–5467. doi: 10.1128/JB.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw LN, Lindholm C, Prajsnar TK, Miller HK, Brown MC, Golonka E, Stewart GC, Tarkowski A, Potempa J. 2008. Identification of sigmaS, a novel component of the Staphylococcus aureus stress and virulence responses. PLoS One 3:e3844. doi: 10.1371/journal.pone.0003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kullik I, Giachino P, Fuchs T. 1998. Deletion of the alternative sigma factor sigmaB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J Bacteriol 180:4814–4820. doi: 10.1128/JB.180.18.4814-4820.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kullik II, Giachino P. 1997. The alternative sigma factor sigmaB in Staphylococcus aureus: regulation of the sigB operon in response to growth phase and heat shock. Arch Microbiol 167:151–159. doi: 10.1007/s002030050428. [DOI] [PubMed] [Google Scholar]
- 7.Pane-Farre J, Jonas B, Hardwick SW, Gronau K, Lewis RJ, Hecker M, Engelmann S. 2009. Role of RsbU in controlling SigB activity in Staphylococcus aureus following alkaline stress. J Bacteriol 191:2561–2573. doi: 10.1128/JB.01514-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giachino P, Engelmann S, Bischoff M. 2001. Sigma(B) activity depends on RsbU in Staphylococcus aureus. J Bacteriol 183:1843–1852. doi: 10.1128/JB.183.6.1843-1852.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pane-Farre J, Jonas B, Forstner K, Engelmann S, Hecker M. 2006. The sigmaB regulon in Staphylococcus aureus and its regulation. Int J Med Microbiol 296:237–258. doi: 10.1016/j.ijmm.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 10.Bischoff M, Entenza JM, Giachino P. 2001. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J Bacteriol 183:5171–5179. doi: 10.1128/JB.183.17.5171-5179.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Homerova D, Bischoff M, Dumolin A, Kormanec J. 2004. Optimization of a two-plasmid system for the identification of promoters recognized by RNA polymerase containing Staphylococcus aureus alternative sigma factor sigmaB. FEMS Microbiol Lett 232:173–179. doi: 10.1016/S0378-1097(04)00063-1. [DOI] [PubMed] [Google Scholar]
- 12.Bischoff M, Dunman P, Kormanec J, Macapagal D, Murphy E, Mounts W, Berger-Bachi B, Projan S. 2004. Microarray-based analysis of the Staphylococcus aureus sigmaB regulon. J Bacteriol 186:4085–4099. doi: 10.1128/JB.186.13.4085-4099.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nair SP, Bischoff M, Senn MM, Berger-Bachi B. 2003. The sigma B regulon influences internalization of Staphylococcus aureus by osteoblasts. Infect Immun 71:4167–4170. doi: 10.1128/IAI.71.7.4167-4170.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S, Reyes D, Beaume M, Francois P, Cheung A. 2014. Contribution of teg49 small RNA in the 5' upstream transcriptional region of sarA to virulence in Staphylococcus aureus. Infect Immun 82:4369–4379. doi: 10.1128/IAI.02002-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nielsen JS, Christiansen MH, Bonde M, Gottschalk S, Frees D, Thomsen LE, Kallipolitis BH. 2011. Searching for small sigmaB-regulated genes in Staphylococcus aureus. Arch Microbiol 193:23–34. doi: 10.1007/s00203-010-0641-1. [DOI] [PubMed] [Google Scholar]
- 16.Morikawa K, Inose Y, Okamura H, Maruyama A, Hayashi H, Takeyasu K, Ohta T. 2003. A new staphylococcal sigma factor in the conserved gene cassette: functional significance and implication for the evolutionary processes. Genes Cells 8:699–712. doi: 10.1046/j.1365-2443.2003.00668.x. [DOI] [PubMed] [Google Scholar]
- 17.Thi Le TN, Romero VM, Morikawa K. 2016. Cell wall-affecting antibiotics modulate natural transformation in SigH-expressing Staphylococcus aureus. J Antibiot (Tokyo) 69:464–466. doi: 10.1038/ja.2015.132. [DOI] [PubMed] [Google Scholar]
- 18.Morikawa K, Takemura AJ, Inose Y, Tsai M, Thi LTN, Ohta T, Msadek T. 2012. Expression of a cryptic secondary sigma factor gene unveils natural competence for DNA transformation in Staphylococcus aureus. PLoS Pathog 8:e1003003. doi: 10.1371/journal.ppat.1003003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen LTT, Takemura AJ, Ohniwa RL, Saito S, Morikawa K. 2018. Sodium polyanethol sulfonate modulates natural transformation of SigH-expressing Staphylococcus aureus. Curr Microbiol 75:499–504. doi: 10.1007/s00284-017-1409-5. [DOI] [PubMed] [Google Scholar]
- 20.Peton V, Breyne K, Rault L, Demeyere K, Berkova N, Meyer E, Even S, Le Loir Y. 2016. Disruption of the sigS gene attenuates the local innate immune response to Staphylococcus aureus in a mouse mastitis model. Vet Microbiol 186:44–51. doi: 10.1016/j.vetmic.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 21.Miller HK, Carroll RK, Burda WN, Krute CN, Davenport JE, Shaw LN. 2012. The extracytoplasmic function sigma factor sigmaS protects against both intracellular and extracytoplasmic stresses in Staphylococcus aureus. J Bacteriol 194:4342–4354. doi: 10.1128/JB.00484-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller HK, Burda WN, Carroll RK, Shaw LN. 2018. Identification of a unique transcriptional architecture for the sigS operon in Staphylococcus aureus. FEMS Microbiol Lett 365:fny108. doi: 10.1093/femsle/fny108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burda WN, Miller HK, Krute CN, Leighton SL, Carroll RK, Shaw LN. 2014. Investigating the genetic regulation of the ECF sigma factor sigmaS in Staphylococcus aureus. BMC Microbiol 14:280. doi: 10.1186/s12866-014-0280-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. 2006. Conserved and variable functions of the sigmaE stress response in related genomes. PLoS Biol 4:e2. doi: 10.1371/journal.pbio.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson KM, Rhodius VA, Gottesman S. 2007. SigmaE regulates and is regulated by a small RNA in Escherichia coli. J Bacteriol 189:4243–4256. doi: 10.1128/JB.00020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Udekwu KI, Wagner EG. 2007. Sigma E controls biogenesis of the antisense RNA MicA. Nucleic Acids Res 35:1018–1037. doi: 10.1093/nar/gkl1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johansen J, Rasmussen AA, Overgaard M, Valentin-Hansen P. 2006. Conserved small non-coding RNAs that belong to the sigmaE regulon: role in down-regulation of outer membrane proteins. J Mol Biol 364:1–8. doi: 10.1016/j.jmb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 28.Johansen J, Eriksen M, Kallipolitis B, Valentin-Hansen P. 2008. Down-regulation of outer membrane proteins by noncoding RNAs: unraveling the cAMP–CRP- and sigmaE-dependent CyaR–ompX regulatory case. J Mol Biol 383:1–9. doi: 10.1016/j.jmb.2008.06.058. [DOI] [PubMed] [Google Scholar]
- 29.Helmann JD. 2016. Bacillus subtilis extracytoplasmic function (ECF) sigma factors and defense of the cell envelope. Curr Opin Microbiol 30:122–132. doi: 10.1016/j.mib.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kingston AW, Liao XB, Helmann JD. 2013. Contributions of the sigmaW, sigmaM and sigmaX regulons to the lantibiotic resistome of Bacillus subtilis. Mol Microbiol 90:502–518. doi: 10.1111/mmi.12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woods EC, Nawrocki KL, Suarez JM, McBride SM. 2016. The Clostridium difficile Dlt pathway is controlled by the extracytoplasmic function sigma factor sigmaV in response to lysozyme. Infect Immun 84:1902–1916. doi: 10.1128/IAI.00207-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho TD, Ellermeier CD. 2011. PrsW is required for colonization, resistance to antimicrobial peptides, and expression of extracytoplasmic function sigma factors in Clostridium difficile. Infect Immun 79:3229–3238. doi: 10.1128/IAI.00019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carroll RK, Weiss A, Broach WH, Wiemels RE, Mogen AB, Rice KC, Shaw LN. 2016. Genome-wide annotation, identification, and global transcriptomic analysis of regulatory or small RNA gene expression in Staphylococcus aureus. mBio 7:e01990-15. doi: 10.1128/mBio.01990-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howden BP, Beaume M, Harrison PF, Hernandez D, Schrenzel J, Seemann T, Francois P, Stinear TP. 2013. Analysis of the small RNA transcriptional response in multidrug-resistant Staphylococcus aureus after antimicrobial exposure. Antimicrob Agents Chemother 57:3864–3874. doi: 10.1128/AAC.00263-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abu-Qatouseh LF, Chinni SV, Seggewiss J, Proctor RA, Brosius J, Rozhdestvensky TS, Peters G, von Eiff C, Becker K. 2010. Identification of differentially expressed small non-protein-coding RNAs in Staphylococcus aureus displaying both the normal and the small-colony variant phenotype. J Mol Med (Berl) 88:565–575. doi: 10.1007/s00109-010-0597-2. [DOI] [PubMed] [Google Scholar]
- 36.Marchais A, Naville M, Bohn C, Bouloc P, Gautheret D. 2009. Single-pass classification of all noncoding sequences in a bacterial genome using phylogenetic profiles. Genome Res 19:1084–1092. doi: 10.1101/gr.089714.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marchais A, Bohn C, Bouloc P, Gautheret D. 2010. RsaOG, a new staphylococcal family of highly transcribed non-coding RNA. RNA Biol 7:116–119. doi: 10.4161/rna.7.2.10925. [DOI] [PubMed] [Google Scholar]
- 38.Anderson KL, Roberts C, Disz T, Vonstein V, Hwang K, Overbeek R, Olson PD, Projan SJ, Dunman PM. 2006. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J Bacteriol 188:6739–6756. doi: 10.1128/JB.00609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pichon C, Felden B. 2005. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc Natl Acad Sci USA 102:14249–14254. doi: 10.1073/pnas.0503838102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beaume M, Hernandez D, Farinelli L, Deluen C, Linder P, Gaspin C, Romby P, Schrenzel J, Francois P. 2010. Cartography of methicillin-resistant S. aureus transcripts: detection, orientation and temporal expression during growth phase and stress conditions. PLoS One 5:e10725. doi: 10.1371/journal.pone.0010725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Battesti A, Bouveret E. 2008. Improvement of bacterial two-hybrid vectors for detection of fusion proteins and transfer to pBAD-tandem affinity purification, calmodulin binding peptide, or 6-histidine tag vectors. Proteomics 8:4768–4771. doi: 10.1002/pmic.200800270. [DOI] [PubMed] [Google Scholar]
- 42.Orr MW, Mao Y, Storz G, Qian S. 2020. Alternative ORFs and small ORFs: shedding light on the dark proteome. Nucleic Acids Res 48:1029–1042. doi: 10.1093/nar/gkz734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown SDM, Lad HV. 2019. The dark genome and pleiotrophy: challenges for precision medicine. Mamm Genome 30:212–216. doi: 10.1007/s00335-019-09813-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hemm MR, Weaver J, Storz G. 2020. Escherichia coli small proteome. EcoSal Plus 9. doi: 10.1128/ecosalplus.ESP-0031-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Storz G, Wolf YI, Ramamurthi KS. 2014. Small proteins can no longer be ignored. Annu Rev Biochem 83:753–777. doi: 10.1146/annurev-biochem-070611-102400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zurek OW, Nygaard TK, Watkins RL, Pallister KB, Torres VJ, Horswill AR, Voyich JM. 2014. The role of innate immunity in promoting SaeR/S-mediated virulence in Staphylococcus aureus. J Innate Immun 6:21–30. doi: 10.1159/000351200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mooney RA, Darst SA, Landick R. 2005. Sigma and RNA polymerase: an on-again, off-again relationship? Mol Cell 20:335–345. doi: 10.1016/j.molcel.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 48.Nystrom T. 2004. Growth versus maintenance: a trade-off dictated by RNA polymerase availability and sigma factor competition? Mol Microbiol 54:855–862. doi: 10.1111/j.1365-2958.2004.04342.x. [DOI] [PubMed] [Google Scholar]
- 49.Missiakas D, Schneewind O. 2013. Growth and laboratory maintenance of Staphylococcus aureus Curr Protoc Microbiol 28:9C.1.1–9C.1.9. doi: 10.1002/9780471729259.mc09c01s28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nair D, Memmi G, Hernandez D, Bard J, Beaume M, Gill S, Francois P, Cheung AL. 2011. Whole-genome sequencing of Staphylococcus aureus strain RN4220, a key laboratory strain used in virulence research, identifies mutations that affect not only virulence factors but also the fitness of the strain. J Bacteriol 193:2332–2335. doi: 10.1128/JB.00027-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kreiswirth BN, Löfdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 52.Olson M. 2015. Bacteriophage transduction in Staphylococcus aureus. Methods Mol Biol 1373:69–74. doi: 10.1007/7651_2014_186. [DOI] [PubMed] [Google Scholar]
- 53.Schneewind O, Missiakas D. 2014. Genetic manipulation of Staphylococcus aureus. Curr Protoc Microbiol 32:Unit 9C.3. doi: 10.1002/9780471729259.mc09c03s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schuster CF, Howard SA, Grundling A. 2019. Use of the counter selectable marker pheS* for genome engineering in Staphylococcus aureus. Microbiology (Reading) 165:572–584. doi: 10.1099/mic.0.000791. [DOI] [PubMed] [Google Scholar]
- 55.Gully D, Bouveret E. 2006. A protein network for phospholipid synthesis uncovered by a variant of the tandem affinity purification method in Escherichia coli. Proteomics 6:282–293. doi: 10.1002/pmic.200500115. [DOI] [PubMed] [Google Scholar]
- 56.Forsyth RA, Haselbeck RJ, Ohlsen KL, Yamamoto RT, Xu H, Trawick JD, Wall D, Wang L, Brown-Driver V, Froelich JM, C KG, King P, McCarthy M, Malone C, Misiner B, Robbins D, Tan Z, Zhu Zy Z-y, Carr G, Mosca DA, Zamudio C, Foulkes JG, Zyskind JW. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol Microbiol 43:1387–1400. doi: 10.1046/j.1365-2958.2002.02832.x. [DOI] [PubMed] [Google Scholar]
- 57.Tomlinson BR, Malof ME, Shaw LN. 2021. A global transcriptomic analysis of Staphylococcus aureus biofilm formation across diverse clonal lineages. Microb Genom 7:e000598. doi: 10.1099/mgen.0.000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S7. Download jb.00392-22-s0001.xlsx, XLSX file, 0.03 MB (32.1KB, xlsx)
Fig. S1 and S2. Download jb.00392-22-s0002.tif, TIF file, 1.6 MB (1.6MB, tif)
Data Availability Statement
Experimental data from this study were deposited in the NCBI Gene Expression Omnibus (GEO) database (GEO accession number GSE215075).