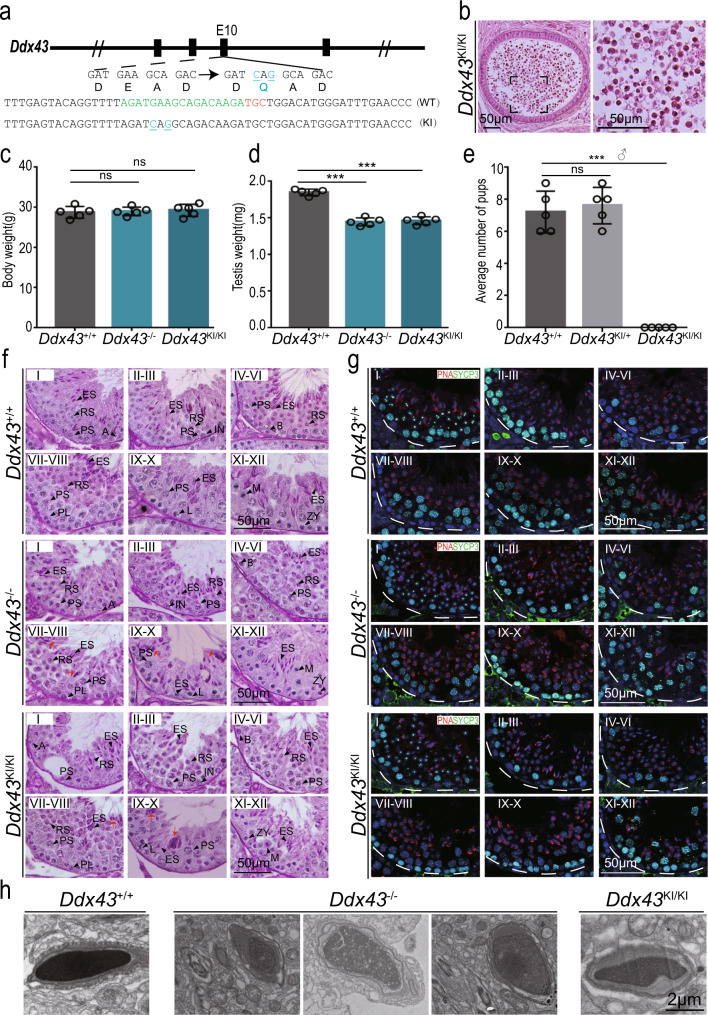
Fig. 2. DDX43 mutant mice exhibit defects in spermatids.
a Using CRISPR-Cas9 to create the ATPase-dead Ddx43 knock-in mice that carry a missense mutation (DEAD to DQAD) in the ATPase motif. Green: sgRNA targeting sequences; blue: mutated sequences; red: PAM nucleotides. b H&E staining on epididymis sections from adult Ddx43KI/KI mice. Ddx43KI/KI epididymis is void of normal spermatozoa that abounds in Ddx43+/+ (Fig.1m), similar to Ddx43–/– (Fig.1n). Scale bars are indicated. c, d Comparative analyses of body weight (c) and testis weight (d) between adult Ddx43+/+, Ddx43-/-, and Ddx43KI/KI mice. N = 5 for each genotype. P values were calculated by oneway ANOVA. ns, not significant; P = 0.9621 (c; Ddx43+/+ vs Ddx43–/–), P = 0.6938 (c; Ddx43+/+ vs Ddx43KI/KI), ***P = 0.0007 (d; Ddx43+/+ vs Ddx43–/–), ***P = 0.0003 (d; Ddx43+/+ vs Ddx43KI/KI). Each bar represents the mean ± SD from biological replicates. e Fertility test on adult Ddx43+/+, Ddx43KI/+, and Ddx43KI/KI mice naturally crossed with age-matched wild-type female mice. N = 5 for each genotype. P values were calculated by oneway ANOVA.ns, not significant; P = 0.8594 (Ddx43+/+ vs Ddx43KI/+), ***P = 0.0006 (Ddx43+/+ vs Ddx43KI/KI). Each bar represents the mean ± SD from biological replicates. f PAS-hematoxylin staining of testis sections from adult Ddx43+/+, Ddx43KI/KI and Ddx43–/– mice. Stages of seminiferous tubule are denoted by Roman numerals. A, type A spermatogonia; IN, intermediate spermatogonia; B, type B spermatogonia; PL, preleptotene spermatocyte; L, leptotene spermatocyte; ZY, zygotene spermatocyte; PS, pachytene spermatocyte; M, metaphase cell; RS, round spermatid; ES, elongating spermatid. Red arrows point to abnormal spermatids at step 16. Scale bars are indicated. g Immunofluorescence staining of SYCP3 (green) and PNA (red) for observation of abnormal spermatids at specific stages of spermatogenesis, parallel to (f). DNA was counterstained with DAPI. Scale bars are indicated. h Representative images of transmission electron microscopy of sperm heads from adult wild-type and mutant mice. Scale bars are indicated. Each experiment was repeated three times with similar results.