Figure 2. NACHT module-containing proteins in bacteria are widespread and diverse.
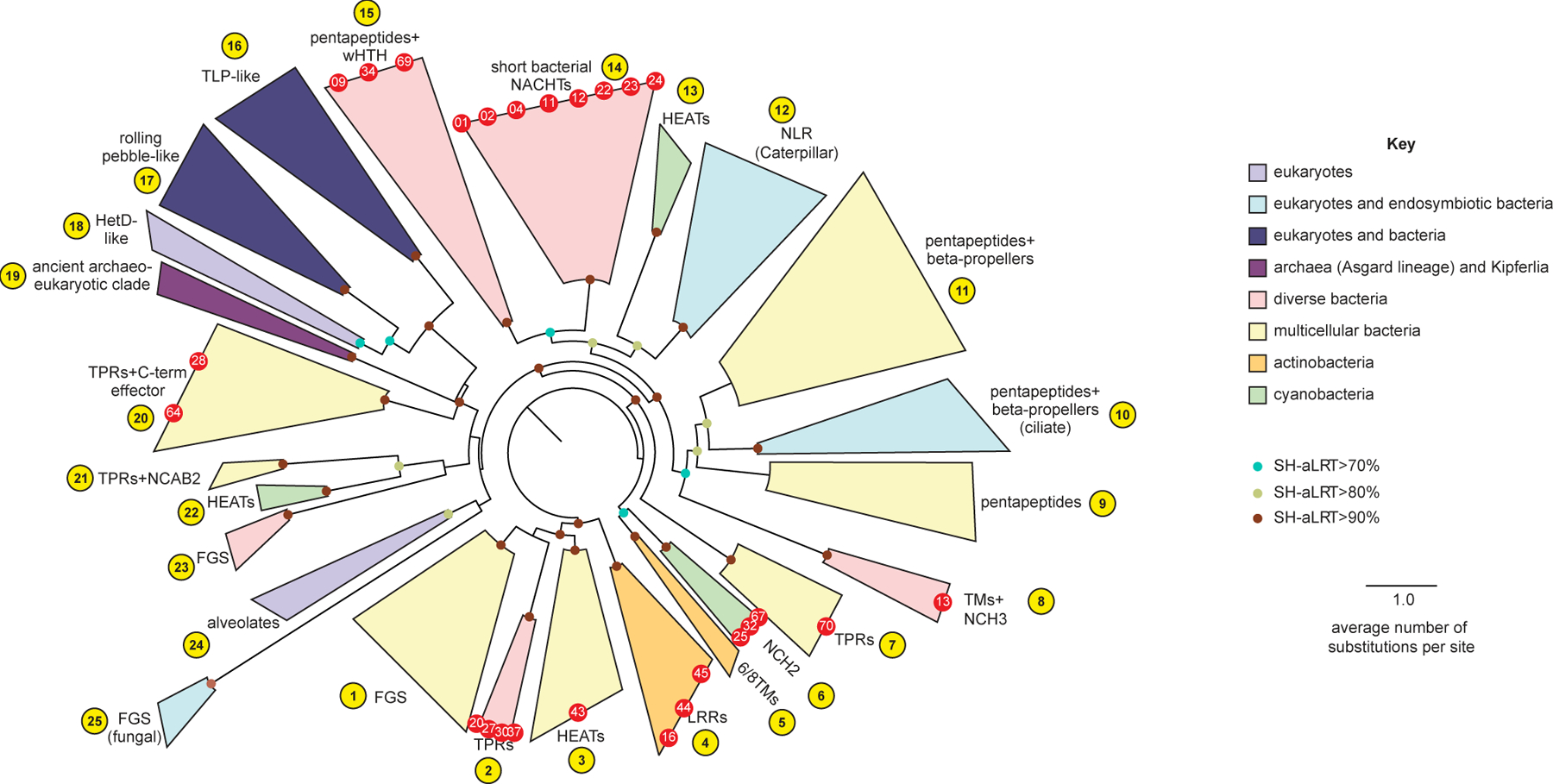
A sequence-based phylogenetic tree of NACHT modules was generated using NACHT module-containing proteins from eukaryotes and prokaryotes. The NACHT module, not accessory domains, were used for tree building. Clades are color-coded based on the indicated key and numbered arbitrarily in yellow circles. Red dots indicate the bacterial NACHT proteins from each clade that were selected for analysis in this study. Bootstrap values are provided where applicable. See Figure S2 for bNACHT gene distribution, Figure S3 for representative domain architectures from each clade, and Table S4 for the most common domain architectures found in each clade. Additional details on genes used to construct the phylogenetic tree can be found in Table S2, and Table S1 contains a full list of all NACHT module-containing proteins identified. See Table S3 for tree topology tests used to validate proposed evolutionary relationships.
