Figure 3. Bacterial NACHT proteins are antiphage.
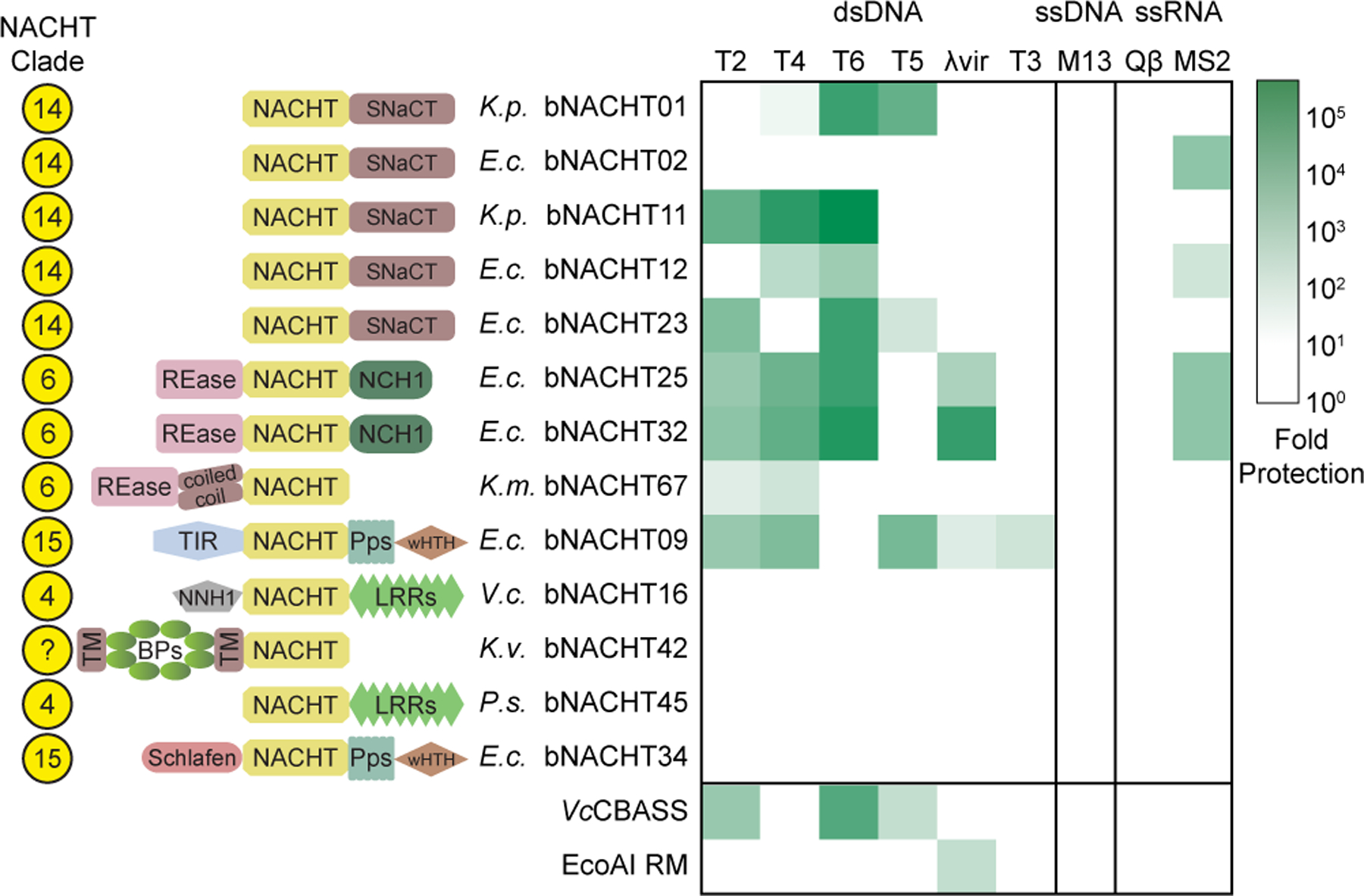
Heat map of fold defense provided by the indicated bNACHT gene for a panel of diverse phages. E. coli expressing the indicated defense system was challenged with phages and fold defense was calculated for each defense system-phage pair by dividing the efficiency of plating (in PFU/mL) on empty vector by efficiency of plating on defense system-expressing bacteria. The NACHT clade, domain architecture, and species of origin for each bNACHT are shown. bNACHT genes displayed in this figure are a subset of the 27 candidates interrogated, selected based on their robust antiphage activity or the diversity of domain architectures sampled. Vibrio cholerae CBASS (VcCBASS) and E. coli UPEC-36 restriction modification system (EcoAI RM) were included as positive controls. Data represent the mean of n = 3 biological replicates. Escherichia coli (E.c.), Klebsiella michiganensis (K.m.), Klebsiella pneumoniae (K.p.), Klebsiella variicola (K.v.), Pseudomonas sp. LAIL14HWK12:I6 (P.s.), Vibrio campbellii (V.c.). Domain abbreviations as described in Figure S3. See Table S5 and Figures S4 and S5 for details on all 27 bNACHT genes analyzed. See Figure S6 for raw efficiency of plating data.
