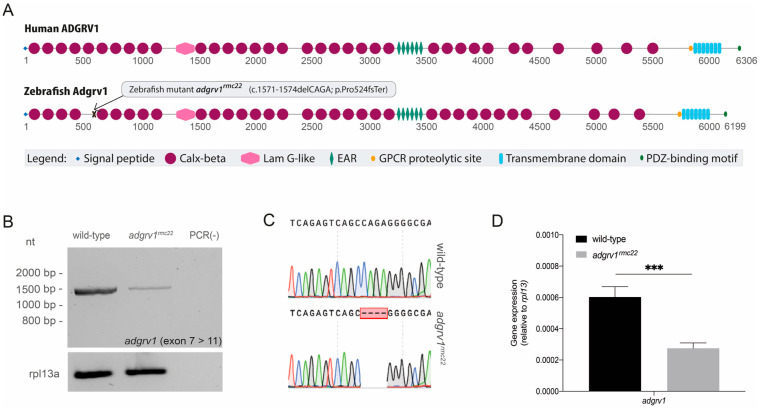
Figure 1.
Domain architecture of human and zebrafish ADGRV1 and transcript analysis in homozygous adgrv1rmc22 mutant zebrafish. (A): Schematic representation of human and zebrafish ADGRV1 protein domain structure. Both proteins have a similar repetitive protein domain architecture. Calx-beta: Ca2+-binding calcium exchanger beta domain; Lam G-like: thrombospondin/pentraxin/laminin G-like domain; EAR: epilepsy-associated repeats; GPCR: G protein-coupled receptor; PDZ: post-synaptic density 95, Discs large, Zonula occludens-1 binding motif. (B): RT-PCR analyses of adgrv1 transcripts derived from homozygous adgrv1rmc22 zebrafish larvae (5 dpf) and wild-type siblings revealed a reduction in mutant adgrv1 transcripts when compared to wild-type adgrv1 transcripts. (C): Sanger sequencing confirmed the four-base-pair deletion in the amplicon derived from homozygous adgrv1rmc22 larvae. (D): RT-qPCR analysis of adgrv1 transcripts in wild-type and adgrv1rmc22 zebrafish larvae (5 dpf). Four pools of five larvae per genotype were used in RT-qPCR analysis. *** indicates p = 0.0001 (two-tailed unpaired Student’s t-test).