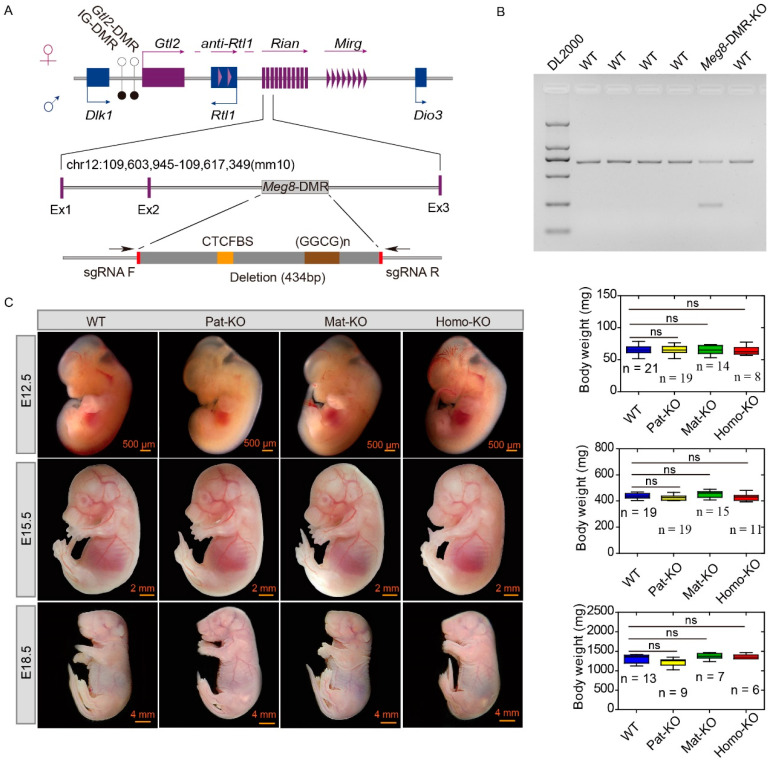
Figure 1.
CRISPR/Cas9-mediated deletion of Meg8-DMR and the growth of the mutants. (A) Regional physical map of the Dlk1-Dio3 domain and deletion locus of Meg8-DMR on mouse chromosome 12. The positions of imprinted genes are indicated by squares, vertical bars (snoRNAs), or triangles (microRNA). Maternally expressed genes are indicated by purple, paternally expressed genes are indicated by blue, CTCFBS is indicated by yellow and (GGCG)n is indicated by brown. The sgRNA primers are indicated by red. The direction of sgRNA primers are indicated by the arrows. IG-DMR and Gtl2-DMR are indicated by circles (filled circles, hypermethylation; open circles, hypomethylation). (B) Demonstration of 434bp deletion in the Meg8-DMR in founder animals. One of the six founders showed a 434bp deletion in the Meg8-DMR. (C) Gross phenotype of E12.5, E15.5, and E18.5 Meg8-DMR embryos. Comparison of WT with Pat-KO, Mat-KO, and Homo-KO mutants.