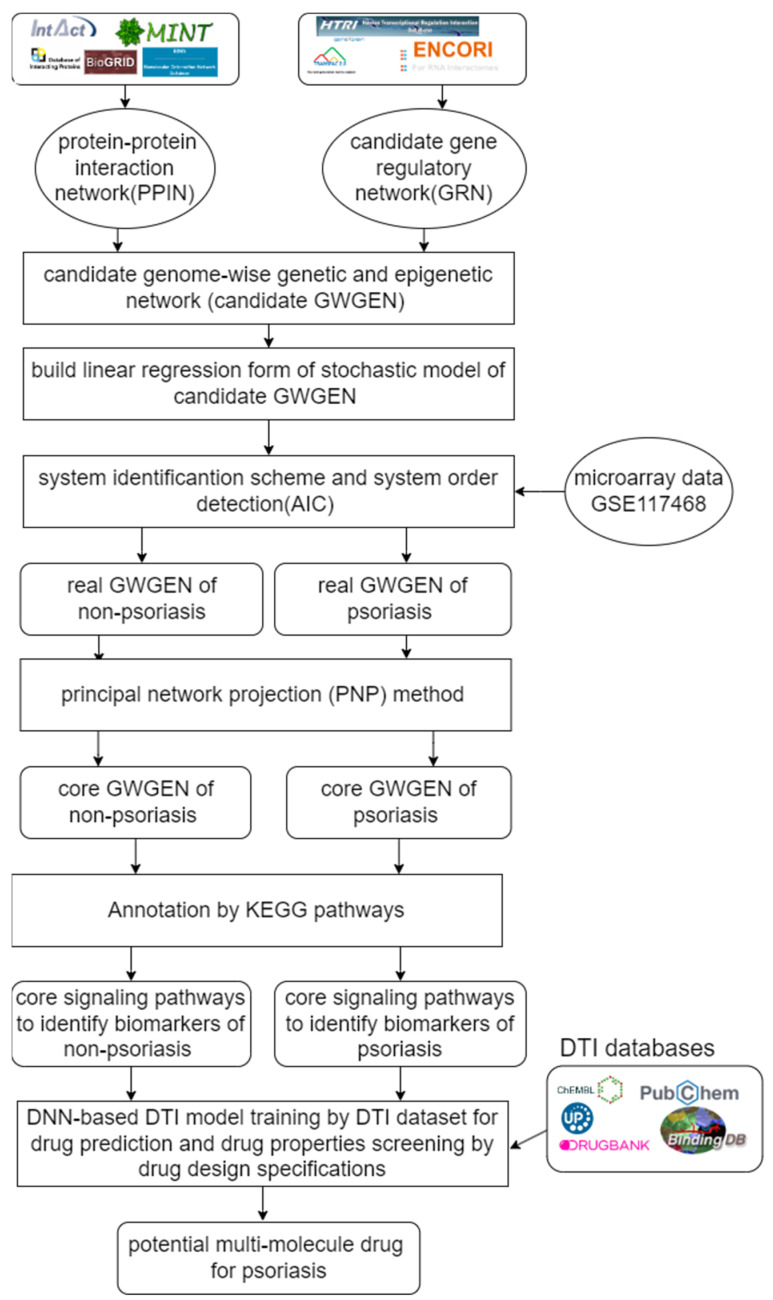
Figure 1.
Flowchart of systems biology methods to identify significant biomarkers of pathogenic mechanism as drug targets of psoriasis and the outline of systematic drug discovery design of psoriasis. Candidate GWGEN is composed of a protein–protein interaction network (PPIN) and gene regulatory network (GRN), which are constructed by PPIN and GRN datasets. Real GWGENs were obtained by pruning false positives from candidate GWGENs by microarray data GSE117468 through system order detection and system identification. The core GWGENs were extracted from real GWGENs by the PNP method. Core GWGENs of psoriasis and non-psoriasis are annotated by KEGG pathways to obtain core signaling pathways of psoriasis and non-psoriasis, respectively. The pathogenic biomarkers were identified for the pathogenesis of psoriasis by comparing the core signaling pathways of psoriasis and non-psoriasis. Finally, the potential drugs were discovered by the prediction of the DNN-based DTI model and the screening of drug design specifications. Then, the screened molecular drugs were combined as a multi-molecule drug for the therapeutic treatment of psoriasis.