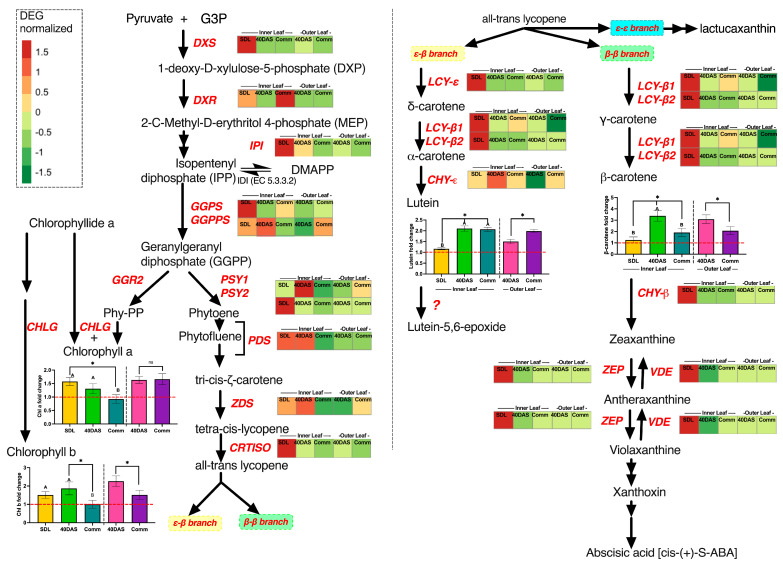
Figure 6.
Metabolomic and transcriptomic pathways of carotenoid biosynthesis in Lactuca sativa, Asteraceae. Genes that encode pathway enzymes are shown in red italic letters. The heatmap is the normalized average of all six varieties at three developmental stages in inner and outer leaves. Abbreviations: GA3P, glyceraldehyde-3-phosphate; DXS, 1-deoxyxylulose 5-phosphate synthase; DOXP, D-1-deoxyxylulose-5-phosphate; DXR, 1-deoxyxylulose 5-phosphate reductoisomerase; MEP, 2-C-methyl-D-erythritol-2,4- cyclodiphosphate; IPP, isopentenyl diphosphate; IPI, IPP isomerase; GGPP, geranylgeranyl diphosphate; GGPS, GGPP synthase; PSY1, phytoene synthase; PDS, phytoene desaturase; ZDS, ζ-carotene desaturase; CRTISO, carotenoid isomerase; LCY-B, lycopene β-cyclase; LCY-E, lycopene ε-cyclase; CHYB, β-ring hydroxylase; CHYE, ε-ring hydroxylase; ZEP, zeaxanthin epoxidase; VDE, violaxanthin deepoxidase; CHLG, Chlorophyll synthase; GGR2, geranylgeranyl reductase2. Bars with similar capital letters at different time point means that samples are not significantly different (p ≥ 0.05), using one-way ANOVA analysis followed by Tukey pos hoc test. * Means that samples are significantly different based on the t-test.