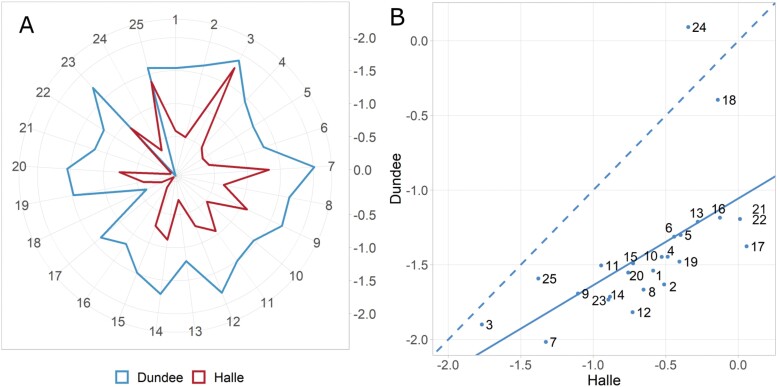
Fig. 1.
Family-specific effect diversity of exotic ELF3 (ELF3Hsp) alleles compared with the cultivated ELF3 (ELF3Hv) alleles for the trait heading in days. Comparison of all 25 families of the barley nested association mapping (NAM) population HEB-25 from Herzig et al. (2018). (A) Each line of the radar plot shows the respective HEB family with its average ELF3Hsp effect on flowering time, compared with the ELF3Hv allele, in days [radar from inside to outside: no acceleration (0) to an acceleration of 2 d (–2)]. (B) Scatterplot comparing ELF3Hsp flowering time effects in days between the two different locations Halle and Dundee. The regression line is shown as a solid line and the dashed line is the diagonal separating effect strengths between the two locations. Pearson’s correlation coefficient between locations is 0.6.