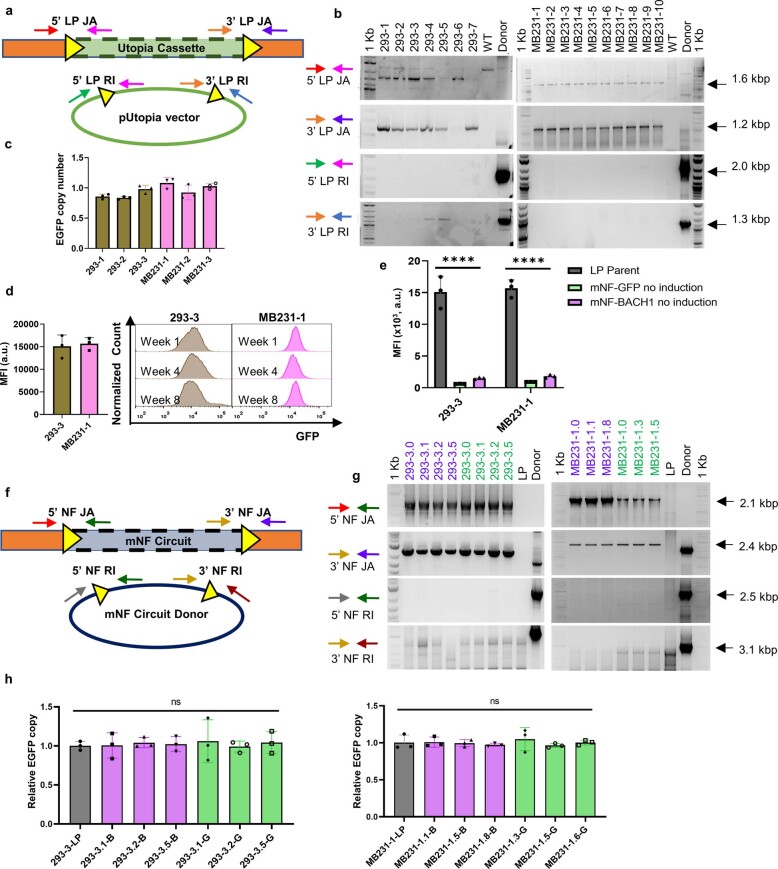
Extended Data Fig. 1. Validation of the integration specificity and quality of Landing Pad and mNF clones.
(a) Strategy to validate the insertion specificity of LP cassette of each monoclonal sample via PCR genotyping. Primers were designed to amplify the adjunction (border) regions between the constructs and the genome at both the (5’ and 3’) ends of the inserts. The adjunction regions with the vector backbones were also tested for any possible random integration. Identical primers are indicated in the same color. (b) PCR genotyping of 293 and MB231 Landing Pad (LP) cell lines using primer sets depicted in (a) for 5’/3’ junction assay (JA) and random integration assay (RI). Parental cells (WT) and Donor plasmid were included as controls. (c) EeGFP copy number determination via qPCR of the selected 293 and MB231 LP cell lines. Each sample copy number was normalized to the reference gene RPPH1 (n = 3). (d) Identical constitutive GFP expression of representative LP cell lines of 293 and MB231 over time (n = 3). (e) Mean fluorescence intensity (MFI) comparison between parental Landing Pad cells, versus uninduced mNF-GFP and mNF-BACH1 integrated cells. n = 3, One-way ANOVA, P < 0.0001. (f) Strategy to validate the insertion specificity of mNF circuits for each monoclonal sample via PCR genotyping. Primers were designed to amplify the adjunction regions between the constructs and the genome at both the (5’ and 3’) ends of the inserts. The adjunction regions with the vector backbones were also tested for any possible random integration. Identical primers are indicated in the same color. (g) PCR genotyping of the mNF-BACH1- (purple) and mNF-GFP- (green) integrated monoclonal populations and polyclonal populations (marked as clone 0) of 293-3 and MB231-1 using primer sets depicted in (f) for 5’/3’ junction assay (JA) and random integration assay (RI). Parental Landing Pad cells (LP) and mNF circuit donor plasmids were included as controls. (h) eGFP copy number determination via qPCR of the selected 293 (left) and MB231 (right) mNF-BACH1- (purple) and mNF-GFP- (green) integrated monoclonal populations relative to the corresponding parental Landing Pad population (LP, grey). Each sample was normalized to corresponding LP sample in each cell type. One-way ANOVA, n = 3.