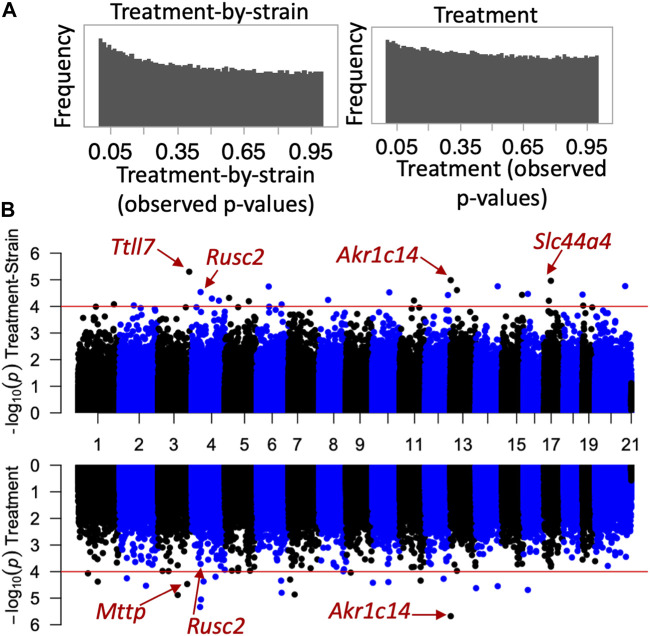
FIGURE 2.
Epigenome-wide association study (EWAS) of CORT+DFP. (A) p-value histograms from the EWAS show a higher frequency of low p-values particularly for the treatment-by-strain interaction effect. This indicates a deviation from the null hypothesis (i.e., a true effect of treatment, but a study that may be underpowered). (B) Mirrored Manhattan plots for treatment-by-strain interaction (top), and treatment effect (bottom). The x-axis plots the location of each CpG region (from chromosomes 1 to X), and the y-axis shows the −Log10P. The red horizontal line marks the nominal significance threshold of −Log10P = 4. Locations of few genes of interest are highlighted.