Figure 3.
Identification of putative enhancers by linking peak to genes
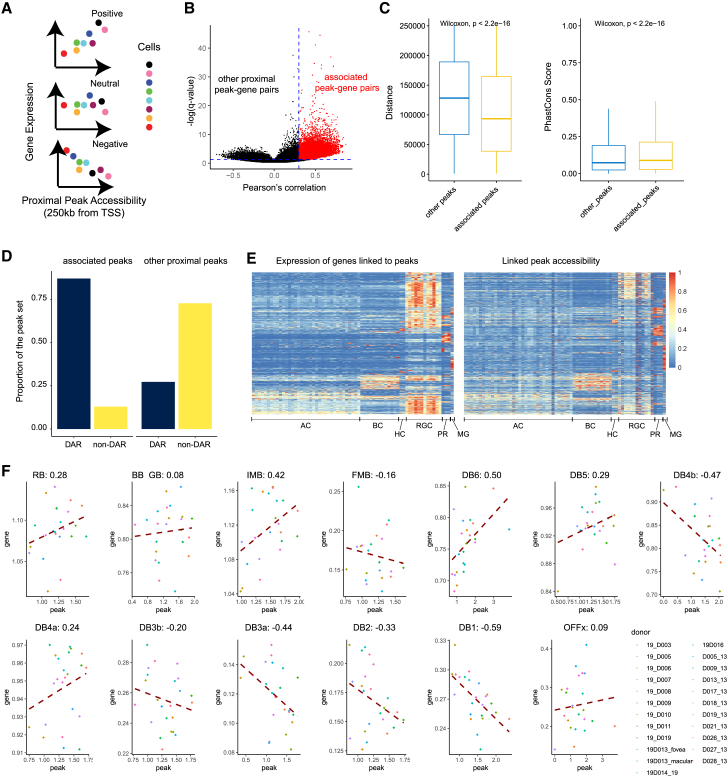
(A) Demonstration of the idea of correlating the gene expression with the proximal peak accessibility. When observing the relationship between a peak and a gene among cells, it is possible that they are positively, negatively, or neutrally correlated.
(B) Volcano plot showing the overall distribution of peak-gene links in two dimensions: their Pearson’s correlation coefficient and negative log-transformed q-values (p values with false discovery rate correction). Red data points highlight the links we considered as putative enhancers.
(C) Boxplot showing the comparison between the putative enhancers and other proximal peaks on their distance to linked gene and sequence conservation. Wilcoxon tests were performed to evaluate the difference between the groups. The center line of the boxplot shows the median of the data; the box limits show the upper and lower quartiles; the whiskers show 1.5 times interquartile ranges.
(D) Bar plot showing the comparison between the putative enhancers and other proximal peaks on their overlapping with major cell class DARs.
(E) Heatmap showing the relative expression of genes linked to peaks and their linked genes across different cell types. The gene expression level and peak accessibility level were both normalized between 0 and 1.
(F) The peak-gene correlation between chr12:6959945-6962406 and GNB3 in all bipolar cell types. For each panel, each data point represents a donor and the red dashed-line represents the linear fit of the correlation between the peak and the gene. The Pearson’s correlation coefficients were labeled on each panel.