Figure 1.
Integrative analysis of the CD69 regulatory landscape
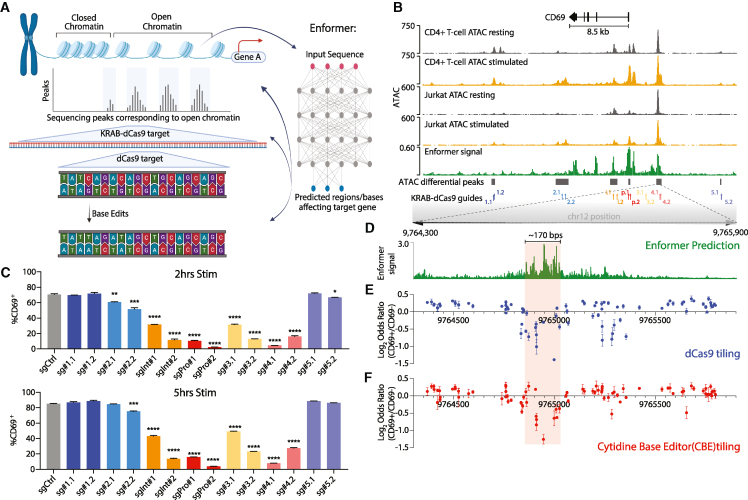
(A) Schematic depicting characterization of the CD69 locus using successive functional perturbations and deep learning.
(B) Genomic tracks depict accessibility of the CD69 locus in primary CD4+ T cells and Jurkat cells, without or with stimulation (PMA/ionomycin). Enformer signal track (summed model gradient) shows the predicted contribution of underlying sequence to CD69 transcriptional output in Jurkat cells. Gray bars depict differentially accessible ATAC peaks in stimulated Jurkat cells relative to resting (FDR < 0.25). CRISPRi sgRNA positions are also indicated. ATAC signal corresponds to reads per genomic content (RPGCs).
(C) Flow cytometry of CD69 expression in Jurkat cells targeted with the indicated CRISPRi sgRNA following a stimulation time course. Samples gated on the live lentiviral transduced population post-puromycin selection.
(D) Expanded view of the absolute value of the Enformer signal (gradient) as described in (B) at single base resolution over RE-4.
(E) Enrichment of dCas9 sgRNAs in CD69+ Jurkat cells relative to CD69− cells (y axis; log2 odds ratio of normalized sgRNA reads). sgRNA positions are plotted along the x axis according to their 5′ starting position on the positive strand. Each data point represents mean ± SEM.
(F) Enrichment/depletion plot of cytidine base editor (CBE) sgRNAs in CD69+ Jurkat cells relative to CD69− cells (as in E). The CBE can edit Cs at base positions 2–11 opposite the NGG PAM, with a strong preference for positions in the central 2–8 base window. sgRNA positions are plotted along the x axis according to their 5′ starting position on the positive strand. Each data point represents mean ± SEM.
For (C), (E), and (F), data represent 2–3 biological independent experiments. A 170 bp region critical for CD69 activation is denoted (D–F, light red).