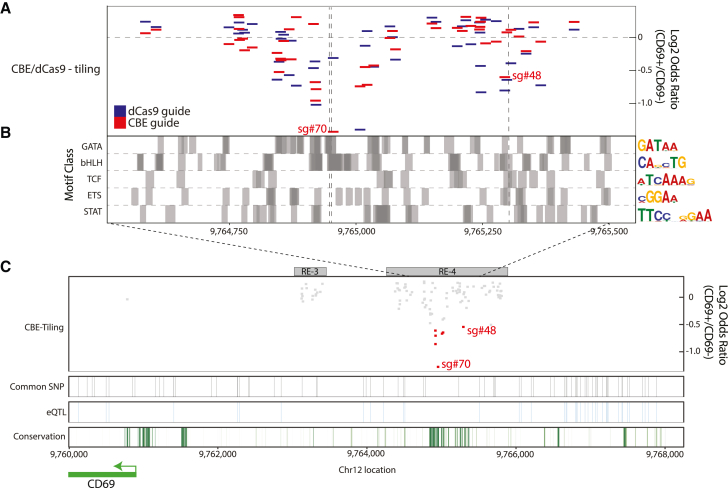
Figure 2.
A critical sequence interval within RE-4 influences CD69 expression
(A) Enrichment and genomic position of sgRNAs in dCas9 and CBE tiling screens as in Figures 1E and 1F limited to the central portion of RE-4. Dashed gray lines correspond to expected C-to-T edit positions for CBE-sgRNAs sg#70 and sg#48.
(B) Transcription factor motif locations (grouped by broad motif class) for key immune regulators shown across the same interval as in (A) (STAR Methods). Dark gray areas represent overlapping motifs. Representative PWM logo plots for each motif class are provided on the right-hand side.
(C) Zoomed-out view of the CD69 locus shows CBE sgRNA depletion (red boxes correspond to top-scoring guides in the sg#70 and sg#48 intervals), common SNPs (black vertical stripes), eQTLs (blue vertical stripes),28 and PhastCon100 conservation score (green stripes).