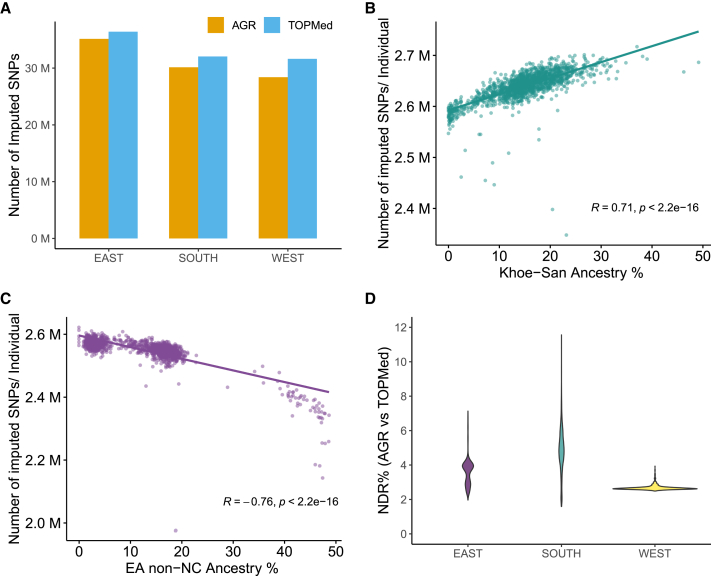
Figure 4.
Impact of geography and non-Niger-Congo ancestry gene flow on imputation
(A) Total number of imputed SNPs in samples from East, West, and South Africa by TOPMed and AGR.
(B) Correlation between the number of SNPs imputed per individual by the AGR and the level of Khoe-San ancestry in South African participants. The regression line, along with correlation coefficient (R) and p value (Pearson correlation), is shown.
(C) Inverse correlation between the number of SNPs imputed per individual by AGR and the level of East African non-Niger-Congo (EA non-NC) ancestry (Afro-Asiatic, Nilo-Saharan, or Eurasian) in the EA participants. The regression line, along with correlation coefficient (R) and p value (Pearson correlation), is shown. The ancestry proportions were inferred using ADMIXTURE (see Figure S5). Ancestry-based variation for the dataset imputed using the TOPMed panel is shown in Figure S6.
(D) Violin plot comparing the distribution of non-reference discordance rate (NDR) between genotypes imputed using AGR vs. TOPMed in the East, West, and South African populations. Each regional subset (i.e., East, West, and South African populations) consist of ∼2,000 participants. The NDR is almost constant across the dataset for West African participants, while the NDR shows substantial variation among the South African participants. Panel codes: AGR, African Genome Resource hosted at the SIS; TOPMed, hosted at the TIS.