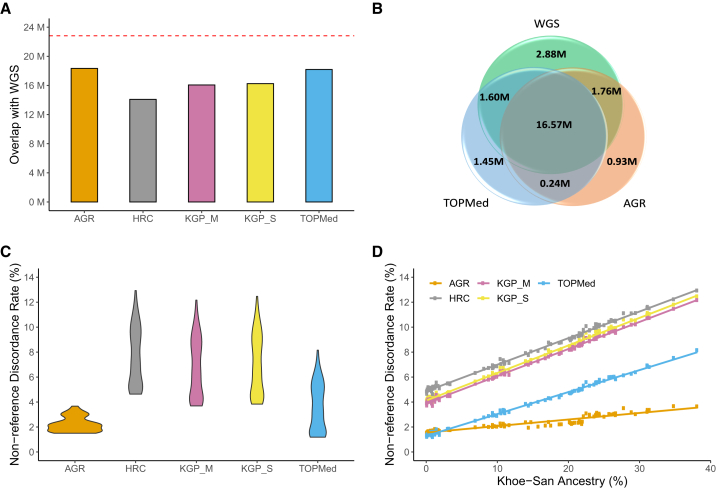
Figure 5.
Comparison of imputed genotypes and genotypes inferred using WGS data
(A) Number of sites that were shared by the imputed and WGS datasets for the 95 individuals. The red line on top shows the number of SNPs in the WGS data.
(B) Venn diagram showing the overlap of SNPs between the WGSs and datasets imputed using AGR and TOPMed panels.
(C) Violin plot summarizing the distribution of NDR for the five panels in the 95 individuals.
(D) Correlation between the overall genotype discordance (estimated by NDR) and the level of Khoe-San ancestry in the five imputed datasets. The regression line for each panel is shown in a different color. The inclusion of the representative Khoe-San population probably leads to a much lower discordance and a gentler slope in the AGR compared with other panels.
AGR, African Genome Resource hosted at the SIS; KGP_S, 1000 Genomes Project hosted at the SIS; HRC, Haplo-type Reference Consortium hosted at the SIS; KGP_M, 1000 Genomes Project hosted at the MIS; TOPMed, hosted at the TIS.