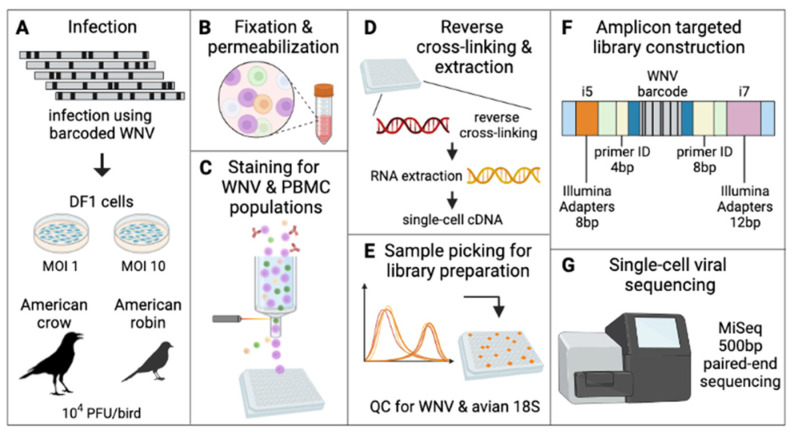
Figure 2.
Approach to barcode quantification of infected DF1 cells and avian PBMCs via flow cytometry and amplicon targeted library construction. (A) DF1 cells at MOI 1 and 10 and American crows and robins at 10,000 PFU/bird were infected with BC-WNV. (B) PBMCs collected from whole avian blood, along with DF1 cells, were separated, fixed and permeabilized. (C) Cells were stained for WNV viral protein and cell type and sorted into a 96-well plate. (D) Cells were reverse cross-linked and viral RNA was extracted and reverse-transcribed into cDNA. (E) Individual cells were classified as WNV infected by qRT-PCR and avian 18S. (F) Libraries were constructed by adapting the Primer ID approach to the Illumina MiSeq platform (Supplementary Materials). (G) Sequencing was performed on an Illumina platform.