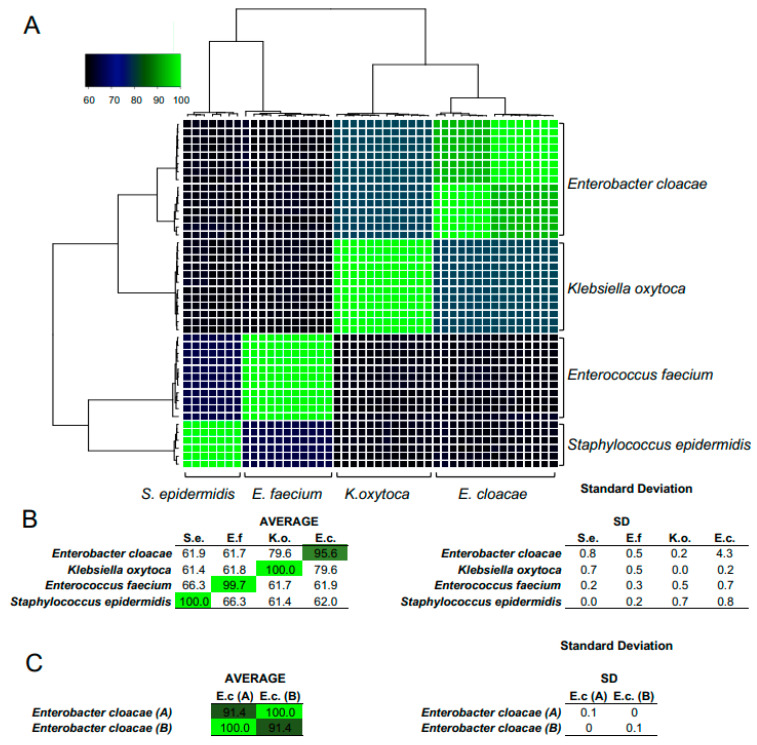
Figure 3.
Whole genome sequencing comparison of bacterial isolates. Heatmap presenting comparison of average nucleotide indices using orthologous fragment pairs (OrthoANI) (A). The total of 47 isolates were subjected to whole genome sequencing (NextSeq, Illumina). SPAdes assembled contiguously were used for classification with kraken2 and for OrthoANI indices comparison. Isolates classified to Kliebsella oxytoca, Enterococcus faecium and Staphylococcus epidermidis showed high-level similarity ranging between 99.7% and 100% (B). Among isolates classified as Enterobacter cloacae, 2 clusters of similarity above 99.9% were identified. The average difference in OrthoANI indices between these 2 clusters reached 9% (C). This could indicate the presence of two Enterobacter cloacae strains within this category.