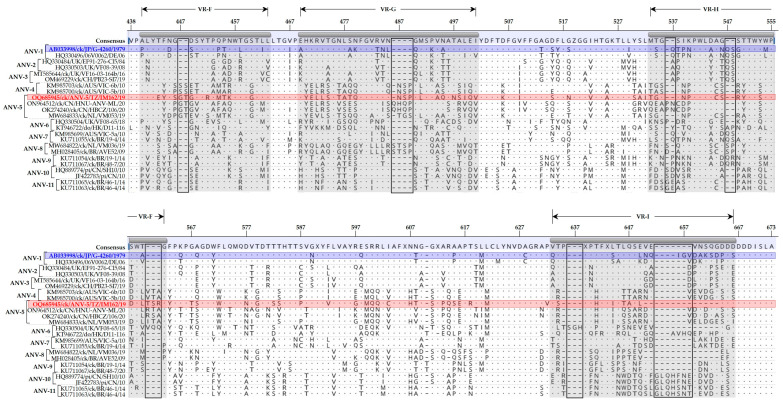
Figure 3.
Variations in the aa residues of the spike (P2) region of the CP sequence of ANV-5/IM162/19 strain (highlighted in red font) compared to representatives of ANV genogroups 1-11 (ANV-1 to ANV-11), shown in square bracket on the left side of the alignment). The aa positions relative to the first (methionine) residue of the protein-coding CP gene are indicated at the top of the consensus (aligned) sequence. The 1979 Japanese ANV-1 strain G-4260 (highlighted in blue), which has been reported to be widely spread in Japanese and European commercial farms [13], is used as reference in the alignment, and four of the nine variable regions (VR-F to VR-I) in the P2 region of the CP sequence are highlighted (shaded in grey color). Identical and missing aa residues are indicated by dots (.) and dashes (-), respectively. Sequence names include GenBank accession numbers, abbreviated host avian species, and country/strain/strain/year of isolation.