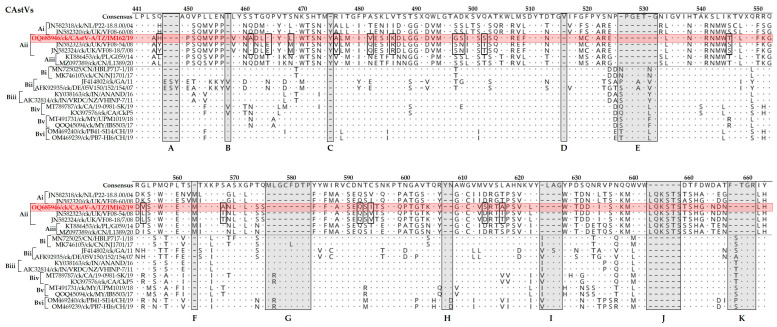
Figure 5.
Variations in the aa residues of the spike (P2) region of the CP sequence of the Tanzanian CAstV-A/IM162/19 strain (highlighted in red) compared to representatives of CAstV subgroups Ai-iii and Bi-Bvi (shown in square bracket on the left side of the alignment). The aa positions relative to the first (methionine) residue of the protein-coding CP gene are indicated at the top of the consensus (aligned) sequence. Dots (.) indicate identical aa residues and dashes (-) indicate missing residues in shaded boxes (labeled A–K). The aa variations between the Tanzanian strain and CAstV-Aii strains from the UK are indicated by open boxes. Sequence names include GenBank accession numbers, abbreviated host avian species of origin, two-letter country abbreviation, strain/strain and year of isolation.