Extended Data Figure 4. The comprehensive profiles of SMARCE1 and SMARCB1 bindings in asynchronous and mitotic ES cells.
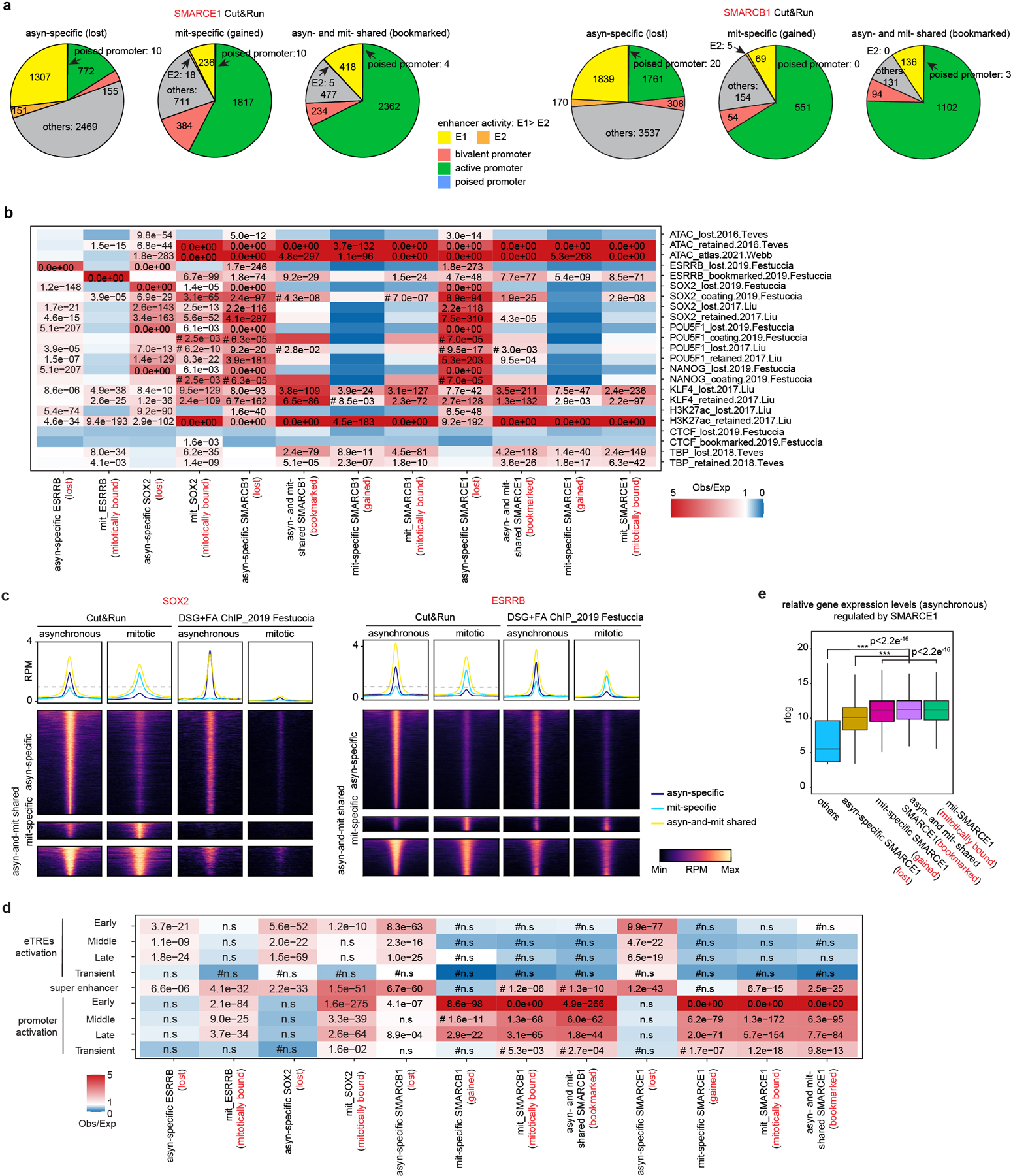
a, The distributions of asyn-specific, mit-specific, asyn- and mit-shared SMARCE1 (left) and SMARCB1 (right) peaks identified from Cut&Run at enhancer and promoter regions in asynchronous and mitotic AB2.2 cells. b, Relative enrichment or depletion of the lost (asynchronous-specific), retained (gained+ bookmarked), gained (mitosis-specific), and bookmarked (present in both asynchronous and mitosis) peaks for ESRRB, SOX2, SMARCB1 and SMARCE1 Cut&Run at the lost, retained, bookmarked, or coating regions of published ATAC signal and indicated factors. Color indicates ratio of observed (Obs) to expected (Exp) frequency, and p-value (two-sided Fisher’s exact test) is indicated if significant (p<0.01, 0.0e00 means p-value<e−350). Comparison using <100 overlapping peaks are denoted with a hash mark (#). c, Average binding profile of Cut&Run and DSG+FA ChIP-seq signal at the indicated binding regions (± 3000-bp peak summit) identified in asynchronous and mitotic mouse ES cells. Average binding profiles represent reads per million (RPM); the y-axis is scaled by median asynchronous binding. d, Relative enrichment or depletion of the lost (asynchronous-specific), retained (gained+ bookmarked), gained (mitosis-specific), and bookmarked (present in both asynchronous and mitosis) peaks for ESRRB, SOX2, SMARCB1 and SMARCE1 Cut&Run at early, middle, late, and transient reactivated enhancer transcriptional regulatory elements (eTREs), super enhancers and promoters. Color indicates ratio of observed (Obs) to expected (Exp) frequency, and p-value (two-sided Fisher’s exact test) is indicated if significant (p<0.01, 0.0e00 means p-value<e−350). Comparison using <100 overlapping peaks are denoted with a hash mark (#). e, Boxplot showing the relative expression levels in asynchronous cells of genes based upon the indicating binding status of SMARCE1. Center lines denote medians; box limits 25th- 75th percentile; whiskers 5th- 95th percentile. Data are compiled from two (a, b, c, d) and eight (e) replicates. Statistical analysis was performed using two-tailed unpaired Student’s t- Test, *** p-value< 0.001 (e).
