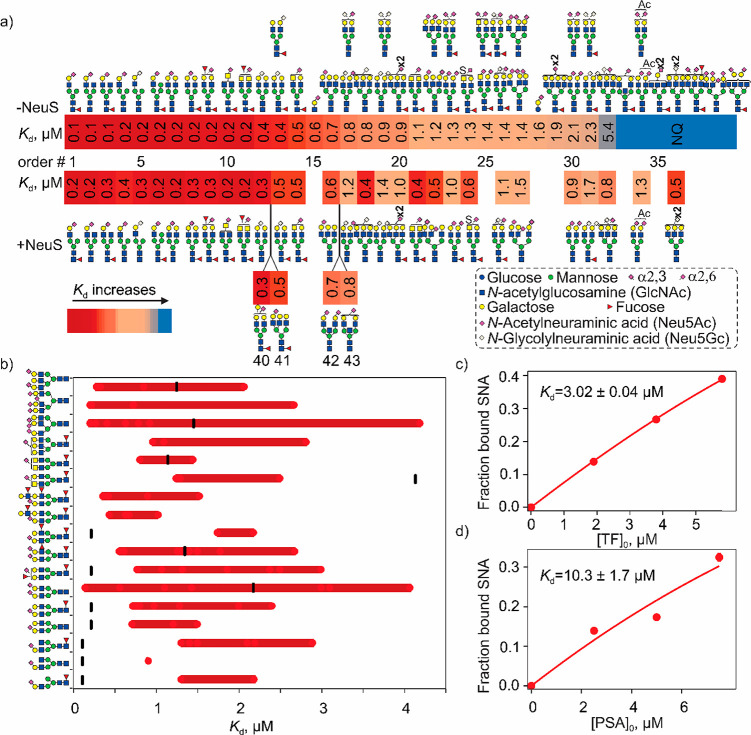
Figure 4.
Glycan affinity rankings measured for natural N-glycan and glycopeptide libraries screened against SNA using COIN-CaR-nMS. (a) Ranking of highest affinity N-glycan ligands detected. When glycans exist as several isoforms, the most abundant one is shown; if several forms are equally abundant, all structures are shown or sialic acid linkage is not specified. The glycan structures from untreated and treated (with neuraminidase) libraries are denoted as −NeuS and +NeuS, respectively. Order number (#) indicates the ranking order. NQ indicates glycan ligand detected but not quantified due to low relative abundance. (b) Range (indicated as red bars) of affinities measured for glycopeptide ligands identified by COIN-CaR-nMS screening. Individual Kd corresponding to different peptide compositions (red circles) and the Kd for the free glycan (black dash) are also shown. (c, d) Concentration-dependent fraction of SNA bound to (c) human transferrin (TF) and (d) prostate cancer antigen (PSA) measured by SLOMO-nMS (SNA 10 μM, PSA 2.5–7.5 μM in Solution 1, 20 μM in Solution 2, TF 2–6 μM in Solution 1, 20 μM in Solution 2). Solid line is the best fit of eq 4a to the data.