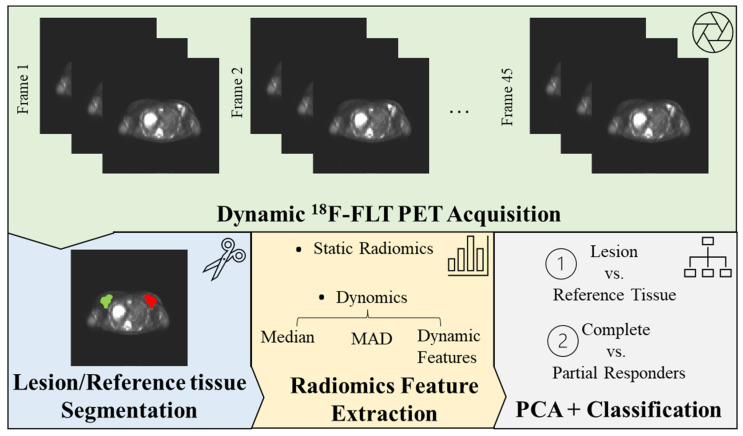
Figure 1.
Radiomics Workflow. Beginning with the acquisition of medical images, volumes of interest (VOIs) are manually segmented on both the lesions and a healthy reference tissue. Radiomic features are subsequently extracted from these VOIs and superimposed onto the static 18F-FLT PET image (a 3D image derived from averaging the final five time-frames of the dynamic acquisition), thus establishing the basis for static radiomics. For our novel dynomics approach, these features are extrapolated from each frame of the dynamic 18F-FLT PET acquisition. In this process, summary values—including median and median absolute deviation (MAD)—are evaluated for each feature and analyzed in conjunction with their temporal evolution (dynamic features). The features encapsulate information about the tumor’s shape, first-order statistical features (derived from the image intensity histogram), and second-order statistical features (texture features). To optimize the data for interpretation, radiomics features undergo redundancy correction via principal component analysis (PCA), enabling the analysis of only non-redundant, meaningful features. These streamlined features are then processed through a machine learning model (XGBoost), generating a clinically interpretable outcome (lesion vs. reference tissue and complete vs. partial responders’ classification).