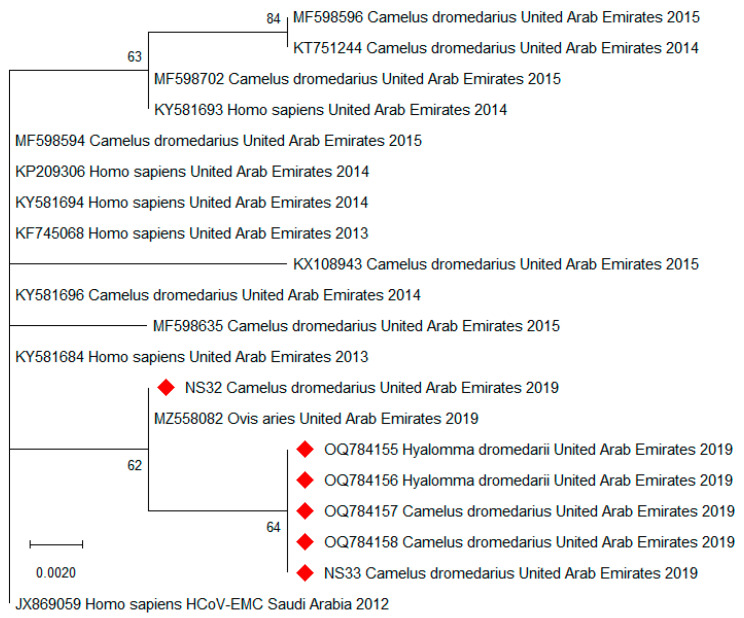
Figure 1.
Maximum likelihood phylogenetic tree of 20 partial MERS-CoV sequences. The 192 bp fragments of the sequences generated in this study (OQ784155-OQ784158, NS32, and NS33; NS = nasal swab; red diamonds) were analyzed together with sequences from humans, camels (Camelus dromedarius), and a sheep (Ovis aries) from the UAE from 2013 to 2019, as well as with the sequence of the first human MERS case detected in Saudi Arabia in 2012 (HCoV-EMC). For each sequence, the corresponding GenBank accession number (where available), host species, country of origin, and collection year are indicated. Horizontal lines represent the genetic distances according to the scale. The tree was inferred using the Jukes–Cantor substitution model over 1000 bootstrap replicates (percentages are displayed at the nodes).