Table 2.
Chemical structures of synthesized oxime derivatives and JNK binding affinity of the compounds.
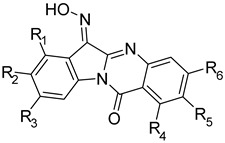
| |||||||||||
| Compd. | R1 | R2 | R3 | R4 | R5 | R6 | JNK1 | JNK2 | JNK3 | Selectivity Index | |
| Kd (µM) | JNK1/JNK3 | JNK2/JNK3 | |||||||||
| TRYP-Ox | H | H | H | H | H | H | 0.15 ± 0.08 | 1.0 ± 0.14 | 0.28 ± 0.21 | 0.15 | 3.6 |
| 4a | H | NO2 | H | H | H | H | N.B. | N.B. | 0.86 ± 0.042 | >40 | >40 |
| 4b | H | Br | H | H | H | H | 3.2 ± 0.21 | 8.5 ± 3.2 | 0.41 ± 0.07 | 7.8 | 20.7 |
| 4c | H | Et | H | H | H | H | 2.6 ± 0.4 | 1.3 ± 0.01 | 0.49 ± 0.09 | 5.3 | 2.7 |
| 4d | H | OMe | H | H | H | H | N.B. | N.B. | 0.34 ± 0.03 | >100 | >100 |
| 4e | H | Ph | H | H | H | H | N.B. | N.B. | 0.34 ± 0.04 | >100 | >100 |
| 4f | H | NO2 | H | H | Br | H | N.B. | N.B. | N.B. | ||
| 4g | Me | H | H | H | H | H | 6.2 ± 1.8 | 14.0 ± 0.7 | 0.40 ± 0.04 | 15.5 | 35 |
| 4h | H | H | Me | H | H | H | 0.67 ± 0.35 | 0.57 ± 0.12 | 0.23 ± 0.04 | 2.9 | 2.5 |
| 4i | H | H | H | H | CF3 | H | N.B. | N.B. | 11.0 ± 1.4 | >3 | >3 |
| 4j | H | H | H | H | Br | H | 4.1 ± 0.6 | 7.4 ± 0.6 | 1.9 ± 0.4 | 2.2 | 3.9 |
| 4k | H | H | H | H | H | Br | N.B. | N.B. | 5.7 ± 1.8 | >6 | >6 |
| 4l | H | H | H | H | H | F | 1.1 ± 0.1 | 0.72 ± 0.22 | 0.43 ± 0.04 | 2.6 | 1.7 |
| 4m | H | H | H | H | H | NO2 | N.B. | N.B. | 6.1 ± 0.4 | >5.6 | >5.6 |
| 4n | H | H | H | H | NO2 | H | N.B. | N.B. | 8.7 ± 1.0 | >3.9 | >3.9 |
| 4o | H | Me | H | H | Me | H | 3.7 ± 0.5 | 6.9 ± 0.2 | 0.87 ± 0.16 | 4.3 | 7.9 |
| 4p | Me | H | H | Me | H | H | N.B. | N.B. | N.B. | ||
| 4q | H | OMe | H | H | OMe | H | N.B. | N.B. | N.B. | ||
| 4r | H | H | OMe | H | H | OMe | 4.6 ± 0.6 | 5.8 ± 1.6 | 3.2 ± 1.1 | 1.4 | 1.8 |
| 4s | H | H | Br | H | H | Br | N.B. | N.B. | N.B. | ||
| 4t | H | Br | H | H | Br | H | N.B. | N.B. | N.B. | ||
| 4u | Br | H | H | Br | H | H | N.B. | N.B. | N.B. | ||
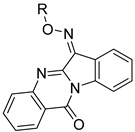
| |||||||||||
| Compd. | R | JNK1 | JNK2 | JNK3 | Selectivity Index | ||||||
| Kd (µM) | JNK1/JNK3 | JNK2/JNK3 | |||||||||
| 5a | -CH2COOEt | N.B. | N.B. | N.B. | |||||||
| 5b | -CH(COOEt)2 | 16.5 ± 0.7 | N.B. | N.B. | |||||||
| 5c | -COOCH3 | 1.7 ± 0.4 | 2.8 ± 0.5 | 0.70 ± 0.12 | 2.4 | 4.0 | |||||
| 5d | -COOCH2-C≡CH | 0.95 ± 0.03 | 1.6 ± 0.1 | 0.32 ± 0.06 | 3.0 | 5.0 | |||||
| 5e |
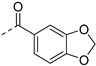
|
0.47 ± 0.08 | 1.5 ± 0.6 | 0.27 ± 0.01 | 1.7 | 0.9 | |||||
N.B., no binding affinity at concentrations < 30 µM.
