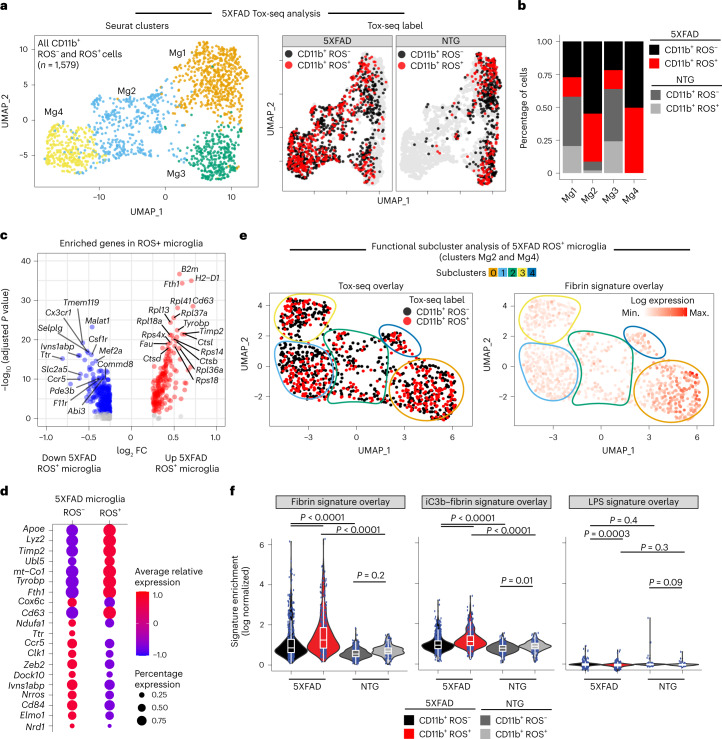
Fig. 6. Single-cell oxidative stress transcriptome of microglia in 5XFAD mice.
a, UMAP plot of single CD11b+ ROS− and CD11b+ ROS+ cells identified by unsupervised clustering analysis (n = 1,579 cells from brains of three 5XFAD and three NTG mice) (left). UMAP of Tox-seq-labeled cells from 5XFAD or NTG mice. b, Fraction of cells in each Seurat cluster colored based on Tox-seq label. c, Volcano plot of enriched DEGs in CD11b+ ROS+ cells from 5XFAD mice. Dots depict average log2 FC and −log10 adjusted P values (log2 FC > 0.25, adjusted P < 0.05 with MAST statistical test with BH correction). d, Dot plot of selected gene markers from c. Average gene expression and cell population expression is depicted as log-normalized scaled expression and percentage, respectively. e, UMAP plots of functional subcluster analysis of 5XFAD CD11b+ ROS+ and CD11b+ ROS− microglia overlaid with Tox-seq label (left) or in vitro fibrin signature (right). f, Violin plots of fibrin, iC3b–fibrin and LPS gene signature overlays with Tox-seq-labeled microglia. Violin plots depict minimum, maximum and median expression, with points showing single-cell expression levels. Box plots show the first to third quartiles (25–75% box bounds) with median values indicated and upper and lower whiskers extending to 1.5× interquartile range. n = 1,579 cells from brains of three 5XFAD and three NTG mice. P < 0.05 as determined by two-way ANOVA with Tukey’s multiple comparison test.