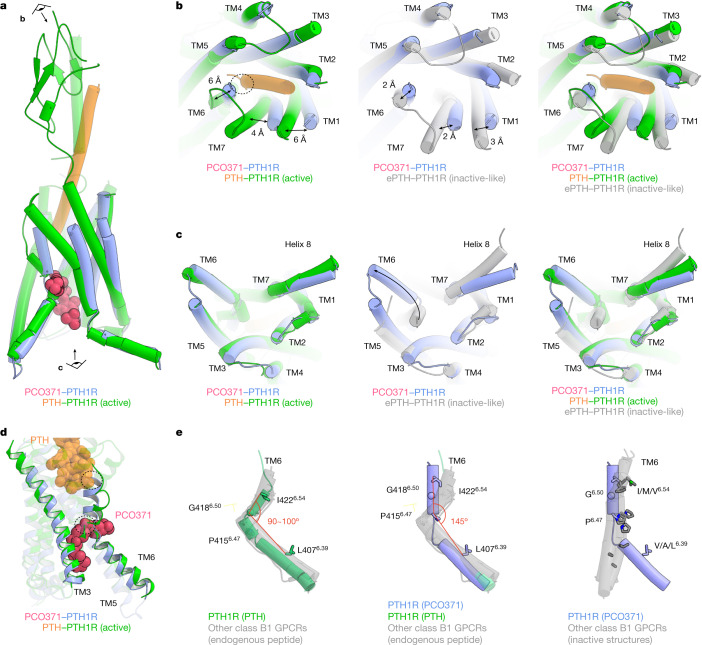
Fig. 2. Comparison of the PCO371-bound PTH1R structure with PTH-bound active PTH1R and ePTH-bound inactive-like PTH1R structures.
a, Superimposition of PCO371–PTH1R–Gs and PTH–PTH1R–Gs (class 2, a representative active form) complexes, aligned on TM2–TM5. The eye and arrow symbols indicate angles of view in b and c. b,c, Extracellular (b) and intracellular (c) views of the superimposed TMDs of PCO371-bound active PTH1R, PTH-bound active PTH1R and ePTH-bound inactive PTH1R. b, Two-way arrows indicate distances of Cα atoms of Thr1921.44 (TM1), Ile4226.54 (TM6) and Met4457.43 (TM7) residues between the structures of PCO371-bound PTH1R and PTH-bound active PTH1R, or between the structures of PCO371-bound and ePTH-bound inactive-like PTH1R. c, The one-way arrow indicates the typical outwards movement of TM6 in PCO371-bound PTH1R and PTH-bound PTH1R. d, Allosteric competitive binding mechanism of PCO371 and PTH. PTH and PCO371 clash at the dashed circles with the other ligand-bound conformation. e, Superimposed structures of PTH-bound PTH1R, PCO371-bound PTH1R, inactive or inactive-like class B1 GPCRs (PTH1R, GCGR, GLP-1R and CRF1R), and all other structures of endogenous agonist-bound class B1 GPCR (PTH2R, SCTR, GHRHR, PAC1R, VIP1R, VIP2R, GCGR, GIPR, GLP-1R, GLP-2R, CALCR, CALRL, CRF1R and CRF2R). The angle is shown among the Cα atoms of Ile/Met/Val6.54, Gly6.50, and Val/Ala/Leu6.39 residues in TM6. Note that the angle of PCO371-bound PTH1R is calculated among Leu6.39, Pro6.47 and Ile6.54 owing to the kink at Pro6.47.