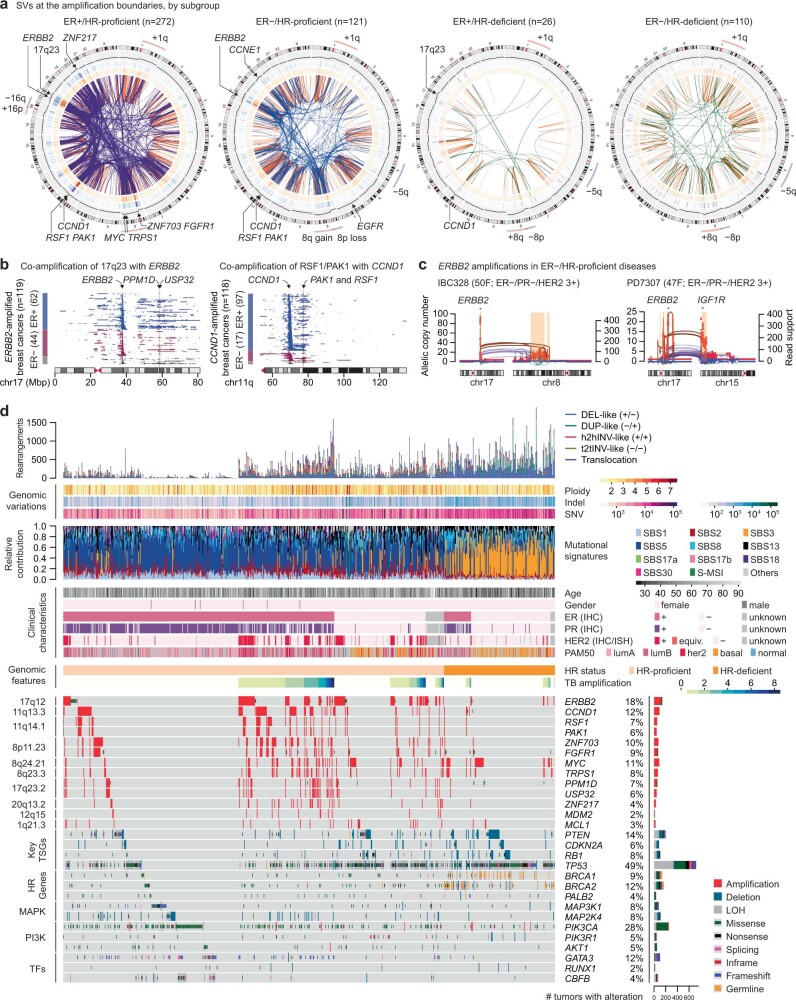
Extended Data Fig. 2. Genomic alterations in 780 breast cancers and their relationship with translocation-bridge amplification.
a. Landscape of boundary SVs by the ER and homologous recombination (HR) status. Frequently amplified genes and common aneuploidies are annotated. b. Amplified genomic segments in ERBB2-amplified tumors by the ER status (left panel). ERBB2 and two oncogenes in 17q23 are annotated. Co-amplification of the 17q23 region (harboring PPM1D, MIR21, USP32, etc.) was numerically more frequent in the HER2+/ER+ group compared to the HER2+/ER− but without statistical significance (37% vs 18%; odds ratio = 2.63; 95% CI = 0.98−7.71 by two-sided Fisher’s exact test). Amplified genomic regions in CCND1-amplified tumors by the ER status (right panel). CCND1, PAK1, and RSF1 are shown as shaded areas. CCND1 amplification was more common in the ER+ group compared to the ER− (21% vs 6%; odds ratio=3.85; 95% CI=2.22−7.05 by two-sided Fisher’s exact test). c. Representative cases of ERBB2 amplification in ER−/HR-proficient breast cancers (ER−/HER2+). The pattern of copy-number amplification with frequent translocations at their boundaries is similar to what was observed in the ER+/HER2+ cases. d. Whole-genome alteration landscape of 780 breast cancer cases shows the relationship between the translocation-bridge amplifications and driver genetic alterations. IHC, immunohistochemical stain; ISH, in situ hybridization; HR, homologous recombination; TSG, tumor suppressor gene; MAPK, mitogen-activated protein kinase; PI3K, phosphatidylinositol-3-kinase; TF, transcription factor.