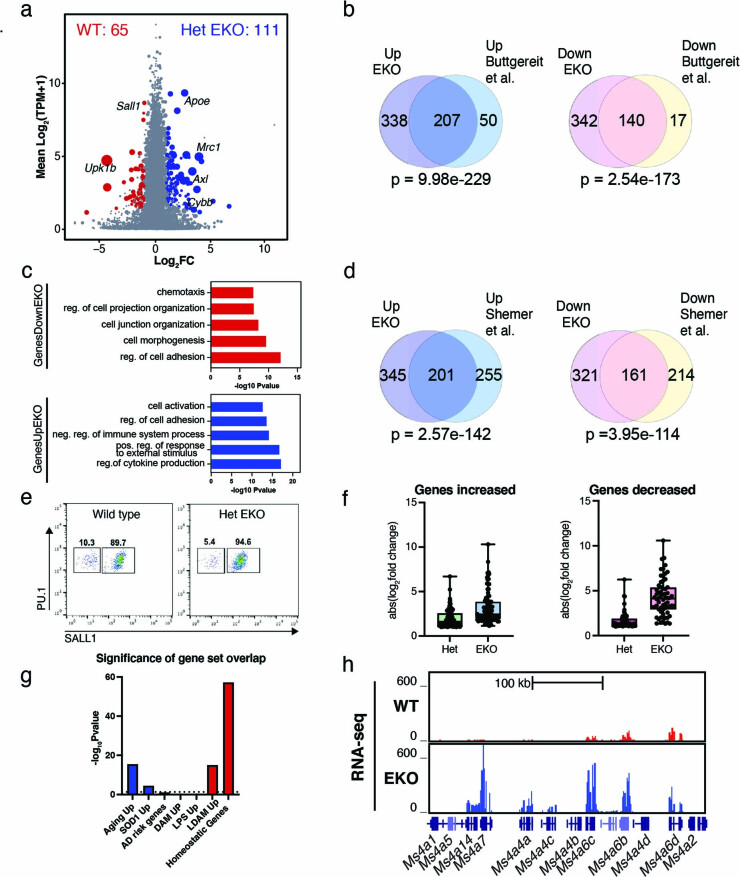
Extended Data Fig. 4. Transcriptional changes in Sall1 EKO and Het EKO microglia.
a. MA plot of RNA-seq data from WT versus Het EKO microglia. n = 3/genotype. b. Overlap of differential genes (p-adj. < 0.05 and log2FC > 1, from DESeq2 analysis (Wald’s test with multiple testing correction using Benjamini–Hochberg method)) identified in EKO microglia vs WT and Sall1 conditional knockout versus control mouse microglia from Buttgereit et al.3. P-values for overlaps were calculated using one-tailed Fisher exact test. c. Metascape GO analysis of pathways significantly changed in EKO vs WT microglia. d. Overlap of differential genes (p-adj. < 0.05 and log2FC > 1, from DESeq2 analysis (Wald’s test with multiple testing correction using Benjamini–Hochberg method)) identified in EKO vs WT microglia and engrafted vs. endogenous microglia (Shemer et al., 2018)25. P-values for overlaps calculated using one-tailed Fisher exact test. e. Flow cytometry of WT and Het EKO brain nuclei stained for PU1 and SALL1. f. Boxplot of log2FC of differential genes shared between Het and EKO microglia (n = 3/genotype). Median (center line), whiskers (max, min), box edges (25th - 75th percentile). g. Significance of gene set overlaps from Fig. 2g. P-values were calculated using one-tailed Fisher exact test for the overlap between differentially expressed genes in the Sall1 EKO vs differentially expressed genes in aging29, the SOD model of ALS29, AD risk genes32, upregulated genes in Disease Associated Microglia (DAM)30, genes upregulated in LPS29, genes upregulated in lipid droplet associated microglia (LDAMs)31 and microglia homeostatic genes10,11,40. Dotted line represents p-value = 0.05. h. Expression of Ms4 family genes in EKO and WT microglia.