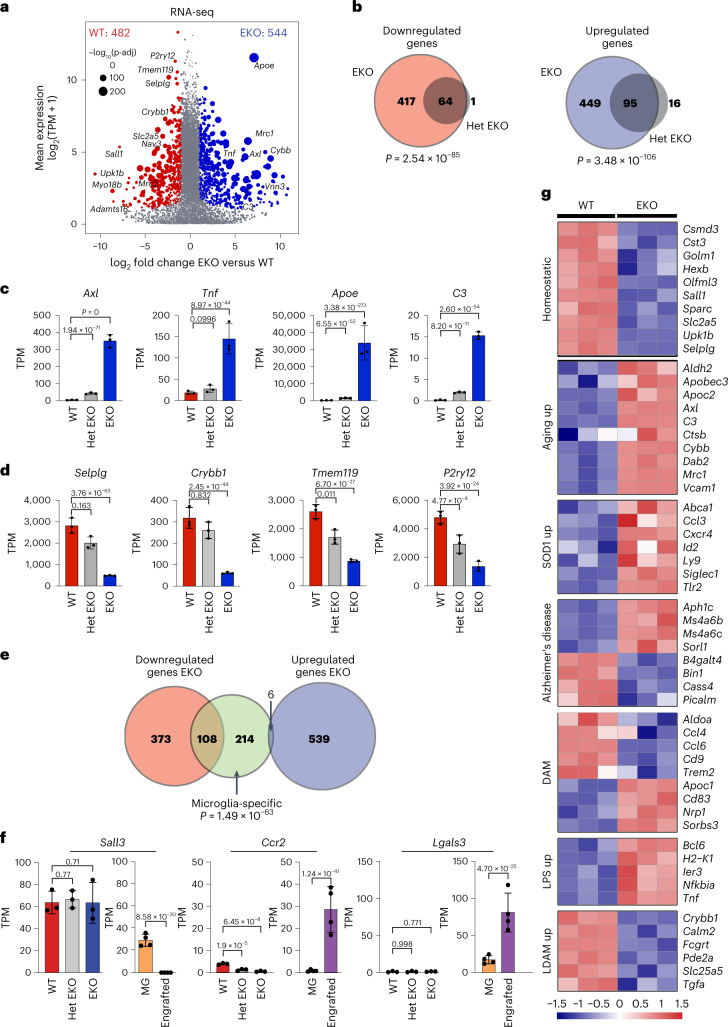
Fig. 2. EKO microglia exhibit a loss of microglia identity and an increased signature of aging and inflammation.
a, MA plot of RNA-seq data comparing WT and EKO microglia. n = 3 per group. DEGs (DESeq2 analysis with Wald’s test with multiple testing correction using Benjamini–Hochberg method) are defined as p-adj <0.05, FC >2 or <−2, and log2(TPM + 1) >2 in at least one group. b, Comparison of overlap between genes increased and decreased in EKO and Het EKO microglia as compared with WT microglia. P values were calculated using one-tailed Fisher exact test. See also Extended Data Fig. 4. c, Bar plots for expression of upregulated genes in WT as compared with Het EKO and EKO microglia. Red, WT; gray, Het EKO; blue, EKO. n = 3 per genotype. Data are represented as mean with standard deviation, p-adj from DESeq2 analysis (Wald’s test with multiple testing correction using Benjamini–Hochberg method) d, Bar plots for expression of downregulated genes in WT as compared with Het EKO and EKO microglia. Red, WT; gray, Het EKO; blue, EKO. n = 3 per genotype. Data are represented as mean with standard deviation, p-adj from DESeq2 analysis (Wald’s test with multiple testing correction using Benjamini–Hochberg method). e, Overlap of significantly downregulated and upregulated genes in EKO versus genes expressed more highly in microglia than other TRMs (Supplementary Table 1). P value for overlaps was calculated using one-tailed Fisher exact test. f, Bar plots for expression of DEGs between resident microglia (MG) and peripherally engrafted microglia-like cells from Shemer et al.25 (n = 4 per group), and in WT, Het EKO and EKO microglia from the present study (n = 3 per genotype). Data are represented as mean with standard deviation, p-adj from DESeq2 analysis (Wald’s test with multiple testing correction using Benjamini–Hochberg method). g, Heat map of DEGs (p-adj from DEseq2 <0.05) in EKO versus WT microglia that are associated with diverse microglia phenotypes (aging29, the SOD model of ALS29, AD risk genes32, DAM30, LPS-treated29, LDAMs31 and homeostatic microglia10,11,39). Each row is z-score-normalized counts for each gene.