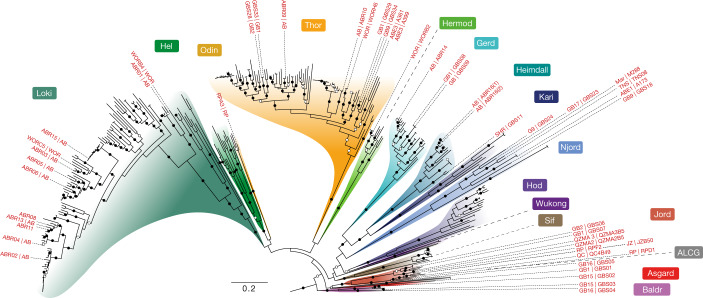
Fig. 1. Phylogenomic analysis of 15 concatenated ribosomal proteins expands Asgard archaea diversity.
ML tree (IQ-TREE, WAG+C60+R4+F+PMSF model) of concatenated protein sequences from at least 5 genes, encoded on a single contig, of a RP15 gene cluster retrieved from publicly available and newly reported Asgard archaeal MAGs. Bootstrap support (100 pseudo-replicates) is indicated by circles at branches, with filled and open circles representing values equal to or larger than 90% and 70% support, respectively. Leaf names indicate the geographical source and isolate name (inner and outer label, respectively) for the MAGs reported in this study. Only the in-group is shown (263 out of 542 total sequences). Scale bar denotes the average number of substitutions per site. AB, Aarhus Bay (Denmark); ABE, ABE vent field, Eastern Lau Spreading Center; ALCG, Asgard Lake Cootharaba Group; Asgard, Asgardarchaeia; Baldr, Baldrarchaeia; GB, Guaymas Basin (Mexico); Gerd, Gerdarchaeales; Hel, Helarchaeales; Heimdall, Heimdallarchaeaceae; Hermod, Hermodarchaeia; Hod, Hodarchaeales; Jord, Jordarchaeia; JZ, Jinze (China); Kari, Kariarchaeaceae; Loki, Lokiarchaeales; Mar, Mariner vent field, Eastern Lau Spreading Center; Njord, Njordarchaeales; Odin, Odinarchaeia; QC, QuCai village (China); QZM, QuZhuoMu village (China); RP, Radiata Pool (New Zealand); SHR, South Hydrate Ridge; Sif, Sifarchaeia; Thor, Thorarchaeia; TNS, Taketomi Island (Japan); WOR: White Oak River (USA); Wukong, Wukongarchaeia.