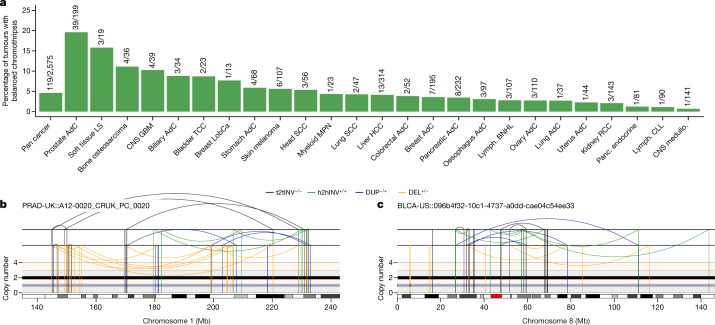
Fig. 5. Prevalence of DNA copy-number-neutral, balanced chromothripsis events across pan-cancer genomes.
a, The frequency of balanced chromothripsis in the ICGC/TCGA PCAWG cohort. The fractions represent the number of tumours with balanced chromothripsis in at least one chromosome over the total number of tumours of each type analysed. AdC, adenocarcinoma; CNS GBM, central nervous system glioblastoma; CNS medullo., central nervous system medulloblastoma; Head SCC, head-and-neck squamous cell carcinoma; HCC, hepatocellular carcinoma; LS, liposarcoma; LobCa, lobular carcinoma; Lymph. BNHL, lymphoid mature B cell lymphoma; Lymph. CLL, lymphoid chronic lymphocytic leukemia; MPN, myeloproliferative neoplasm; Panc. endocrine, pancreatic neuroendocrine tumour; RCC, renal cell carcinoma; TCC, transitional cell carcinoma. b,c, Examples of balanced chromothripsis events in prostate adenocarcinoma (b) and bladder cancer (c) characterized by clusters of interleaved rearrangements, as expected for the random rejoining of genomic fragments shattered in chromothripsis, but without DNA loss, as indicated by the lack of deletions. The total and minor copy-number data are represented in black and grey, respectively. DEL, deletion-like rearrangement; DUP, duplication-like rearrangement; h2hINV, head-to-head inversion; t2tINV, tail-to-tail inversion. An example of canonical chromothripsis and additional examples of balanced chromothripsis events are provided in Extended Data Fig. 10.